-
Notifications
You must be signed in to change notification settings - Fork 11
Home
This wiki is obsolete, please visit the documentation here https://uammd.readthedocs.io/en/latest/ instead.
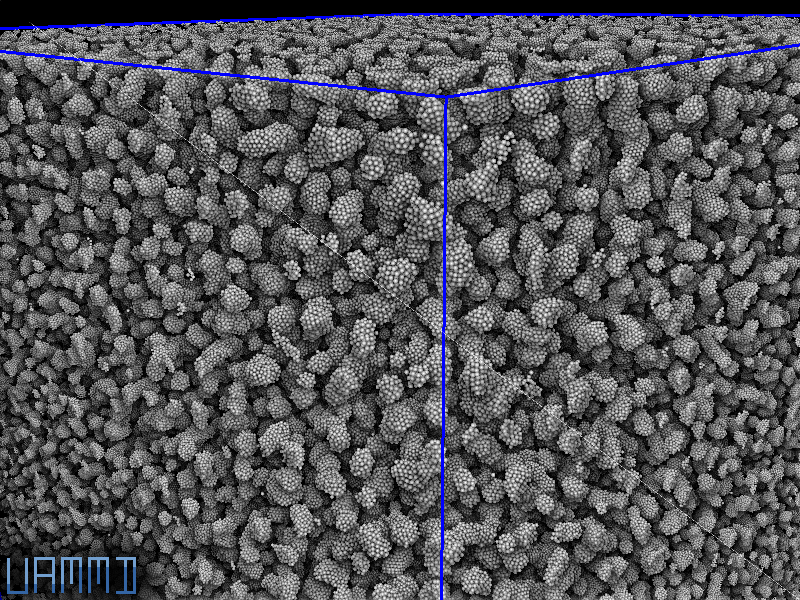
10 Million LJ particles in a box with a low temperature thermostat. Ran on a GTX 980.
Visualization is made with superpunto! https://github.com/RaulPPelaez/superpunto/
For information about a particular module, see its wiki page. You will find the most up to date information about a module in its header file.
UAMMD is a CUDA/C++14 header-only code.
Compiling and running any code containing UAMMD requires a GPU and a system with a working CUDA environment. CUDA is free and you can download it here:
https://developer.nvidia.com/cuda-toolkit
On the other hand understanding and modifying UAMMD code will require at least basic knowledge of GPU programming with CUDA. There are a lot of resources online to learn CUDA, but you can start here:
https://developer.nvidia.com/blog/even-easier-introduction-cuda/
If this is the first time you encounter UAMMD and want a quick start guide check the Quick Start wiki page.
Look in Compiling UAMMD if some Makefile gives you troubles. The FAQ is also a good source of knowledge.
UAMMD is a header only framework and it is mostly self contained in terms of dependencies so compilation should not be too troubling.
-
-
1. PairForces
2. NbodyForces
3. ExternalForces
4. BondedForces
5. AngularBondedForces
6. TorsionalBondedForces
7. Poisson (Electrostatics) -
-
MD (Molecular Dynamics)
1. VerletNVT
2. VerletNVE - BD Brownian Dynamics
-
BDHI Brownian Dynamics with Hydrodynamic Interactions
1. EulerMaruyama
1.1 BDHI_Cholesky Brownian displacements through Cholesky factorization.
1.2 BDHI_Lanczos Brownian displacements through Lanczos algorithm.
1.3 BDHI_PSE Positively Split Edwald.
1.4 BDHI_FCM Force Coupling Method. - DPD Dissipative Particle Dynamics
- SPH Smoothed Particle Hydrodynamics
-
Hydrodynamics
1. ICM Inertial Coupling Method.
2. FIB Fluctuating Immerse Boundary.
3. Quasi2D Quasi2D hydrodynamics
-
MD (Molecular Dynamics)
-
- 1. Neighbour Lists
-
1. Programming Tools
2. Utils
-
1. Transverser
2. Functor
3. Potential
-
1. Particle Data
2. Particle Group
3. System
4. Parameter updatable
-
1. Tabulated Function
2. Postprocessing tools
3. InputFile
4. Tests
5. Allocator
6. Temporary memory
7. Immersed Boundary (IBM)
-
1. NBody
2. Neighbour Lists
3. Python wrappers
- 1. Superpunto