-
Notifications
You must be signed in to change notification settings - Fork 44
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
Merge pull request #326 from usnistgov/develop
Develop
- Loading branch information
Showing
15 changed files
with
159 additions
and
25 deletions.
There are no files selected for viewing
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,104 @@ | ||
| # Guide to JARVIS-Leaderboard | ||
|
|
||
| ## Introduction | ||
| JARVIS-Leaderboard is an open-source, community-driven platform that facilitates benchmarking and enhances reproducibility in materials design. Users can set up benchmarks with custom tasks and contribute datasets, code, and meta-data. The platform covers five main categories: Artificial Intelligence (AI), Electronic Structure (ES), Force-fields (FF), Quantum Computation (QC), and Experiments (EXP). | ||
|
|
||
| ## External Resources | ||
| - [Powerpoint slides](https://lnkd.in/eNg4w6Cz) | ||
| - [YouTube video](https://www.youtube.com/embed/QDx3jSIwpMo?autoplay=1&mute=1) | ||
|
|
||
| ## Terminologies | ||
|
|
||
| ### Categories | ||
| - **AI:** Input data types include atomic structures, images, spectra, and text. | ||
| - **ES:** Involves various ES approaches, software packages, pseudopotentials, materials, and properties, comparing results to experiments. | ||
| - **FF:** Focuses on multiple approaches for material property predictions. | ||
| - **QC:** Benchmarks Hamiltonian simulations using various quantum algorithms and circuits. | ||
| - **EXP:** Utilizes inter-laboratory approaches to establish benchmarks. | ||
|
|
||
| ### Sub-categories | ||
| 1. SinglePropertyPrediction | ||
| 2. SinglePropertyClass | ||
| 3. ImageClass | ||
| 4. textClass | ||
| 5. MLFF (machine learning force-field) | ||
| 6. Spectra | ||
| 7. EigenSolver | ||
|
|
||
| ### Benchmarks | ||
| Ground truth data used to calculate metrics for specific tasks (e.g., a json.zip file). | ||
|
|
||
| ### Methods | ||
| Precise specifications for evaluation against a benchmark (e.g., DFT with VASP-GGA-PAW-PBE in the ES category). | ||
|
|
||
| ### Contributions | ||
| Individual data in the form of csv.zip files for each benchmark and method. Each contribution includes: | ||
| - Method (e.g., AI) | ||
| - Category (e.g., SinglePropertyPrediction) | ||
| - Property (e.g., formation energy) | ||
| - Dataset (e.g., dft_3d) | ||
| - Data-split (e.g., test) | ||
| - Metric (e.g., mae) | ||
|
|
||
| ## Directory and File Structure | ||
| 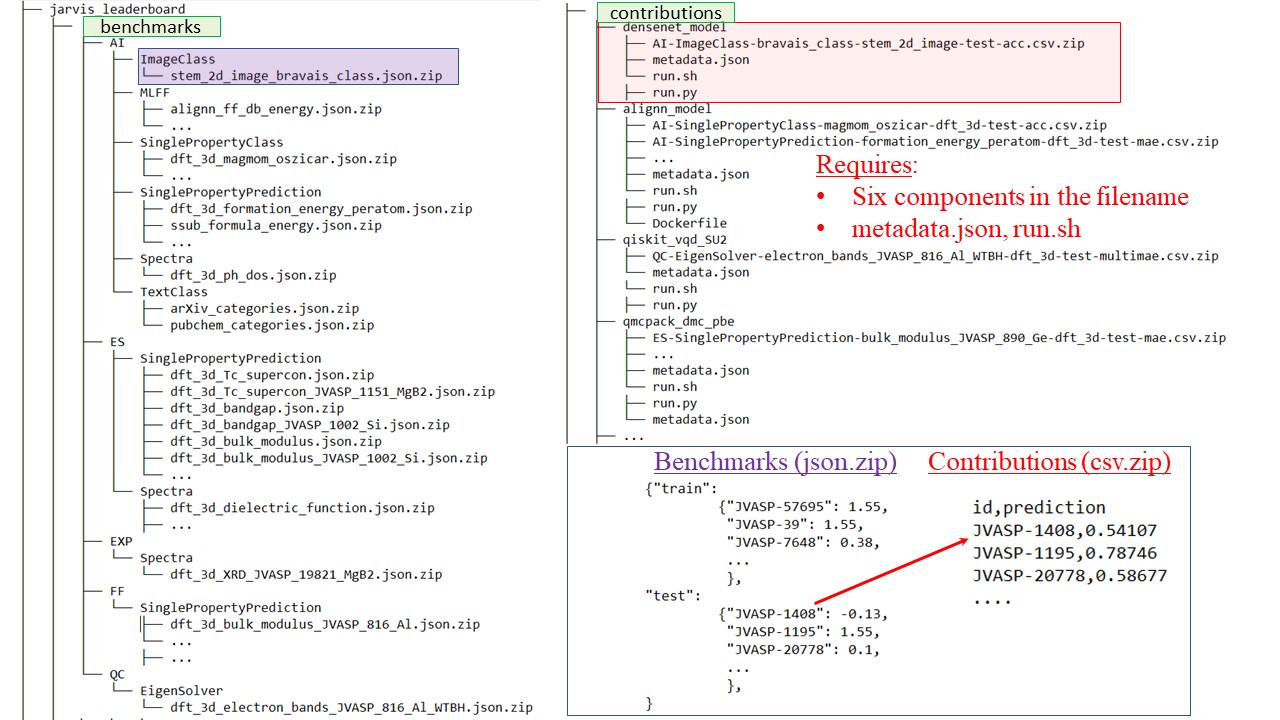 | ||
|
|
||
| ## How to Contribute | ||
|
|
||
| ### Adding a Contribution (csv.zip) | ||
| 1. Fork the JARVIS-Leaderboard repository on GitHub. | ||
| 2. Clone your forked repository: | ||
| ```bash | ||
| git clone https://github.com/USERNAME/jarvis_leaderboard | ||
| ``` | ||
| 3. Create a Python environment: | ||
| ```bash | ||
| conda create --name leaderboard python=3.8 | ||
| source activate leaderboard | ||
| ``` | ||
| 4. Install the package: | ||
| ```bash | ||
| python setup.py develop | ||
| ``` | ||
| 5. Add a contribution: | ||
| ```bash | ||
| cd jarvis_leaderboard/contributions/ | ||
| mkdir vasp_pbe_teamX | ||
| cd vasp_pbe_teamX | ||
| cp ../vasp_optb88vdw/ES-SinglePropertyPrediction-bandgap_JVASP_1002_Si-dft_3d-test-mae.csv.zip . | ||
| vi ES-SinglePropertyPrediction-bandgap_JVASP_1002_Si-dft_3d-test-mae.csv.zip | ||
| ``` | ||
| 6. Modify the prediction value in the csv file, add `metadata.json` and `run.sh` files. | ||
| 7. Rebuild the leaderboard: | ||
| ```bash | ||
| cd ../../../ | ||
| python jarvis_leaderboard/rebuild.py | ||
| mkdocs serve | ||
| ``` | ||
| 8. Commit and push your changes: | ||
| ```bash | ||
| git add jarvis_leaderboard/contributions/vasp_pbe_teamX | ||
| git commit -m 'Adding my PBE Si result.' | ||
| git push | ||
| ``` | ||
| 9. Create a pull request on GitHub. | ||
|
|
||
| ### Adding a New Benchmark (json.zip) | ||
| 1. Create a `json.zip` file in the `jarvis_leaderboard/benchmarks` folder. | ||
| 2. Add a `.json` file with `train`, `val`, `test` keys. | ||
| 3. Add a corresponding `.md` file in the `jarvis_leaderboard/docs` folder. | ||
| 4. Follow instructions for "Adding model benchmarks to existing dataset". | ||
|
|
||
| ## Acronyms | ||
| 1. MAE: Mean Absolute Error | ||
| 2. ACC: Classification accuracy | ||
| 3. MULTIMAE: MAE sum of multiple entries, Euclidean distance | ||
|
|
||
| ## Help | ||
| For questions or concerns, raise an [issue on GitHub](https://github.com/usnistgov/jarvis_leaderboard/issues) or email Kamal Choudhary ([email protected]). | ||
|
|
||
| ## Citation | ||
| [JARVIS-Leaderboard: a large scale benchmark of materials design methods](https://www.nature.com/articles/s41524-024-01259-w) | ||
|
|
||
| ## License | ||
| This template is served under the NIST license. Read the [LICENSE] file for more info. |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
Binary file added
BIN
+670 Bytes
...ontributions/alignn_model/AI-SinglePropertyPrediction-Tc_supercon-dft_3d-test-mae.csv.zip
Binary file not shown.
Binary file added
BIN
+6.53 KB
...s_leaderboard/contributions/atomgpt_model/AI-AtomGen-Tc_supercon-dft_3d-test-rmse.csv.zip
Binary file not shown.
Binary file added
BIN
+273 KB
...erboard/contributions/atomgpt_model/AI-AtomGen-energy_per_atom-carbon24-test-rmse.csv.zip
Binary file not shown.
Binary file added
BIN
+588 Bytes
...ntributions/atomgpt_model/AI-SinglePropertyPrediction-Tc_supercon-dft_3d-test-mae.csv.zip
Binary file not shown.
Binary file added
BIN
+45.7 KB
...tomgpt_model/AI-SinglePropertyPrediction-formation_energy_peratom-dft_3d-test-mae.csv.zip
Binary file not shown.
Binary file added
BIN
+11.8 KB
...ntributions/atomgpt_model/AI-SinglePropertyPrediction-mbj_bandgap-dft_3d-test-mae.csv.zip
Binary file not shown.
Binary file added
BIN
+30.9 KB
...tions/atomgpt_model/AI-SinglePropertyPrediction-optb88vdw_bandgap-dft_3d-test-mae.csv.zip
Binary file not shown.
20 changes: 20 additions & 0 deletions
20
jarvis_leaderboard/contributions/atomgpt_model/metadata.json
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,20 @@ | ||
| { | ||
| "model_name": "AtomGPT", | ||
| "project_url": "https://arxiv.org/abs/2405.03680", | ||
| "date_submitted": "05-23-2024", | ||
| "author_email": "[email protected]", | ||
| "database_version": "12-12-2022", | ||
| "team_name": "AtomGPT", | ||
| "time_taken_seconds": { | ||
| "AI-SinglePropertyClass-mbj_bandgap-dft_3d-test-acc.csv.zip": "", | ||
| "AI-SinglePropertyClass-optb88vdw_bandgap-dft_3d-test-acc.csv.zip": "", | ||
| "AI-SinglePropertyPrediction-formation_energy_peratom-dft_3d-test-mae.csv.zip": "" | ||
| }, | ||
| "language": "python", | ||
| "os": "linux", | ||
| "software_used": "jarvis-tools,numpy,scipy,torch,atomgpt", | ||
| "hardware_used": "nisaba-cluster at NIST, V100 Tesla GPU", | ||
| "git_url": [ | ||
| "https://github.com/usnistgov/atomgpt" | ||
| ] | ||
| } |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,3 @@ | ||
| #!/bin/bash | ||
| pip install atomgpt | ||
| python run.py |
Binary file added
BIN
+717 Bytes
...rboard/contributions/cfid/AI-SinglePropertyPrediction-Tc_supercon-dft_3d-test-mae.csv.zip
Binary file not shown.
Binary file added
BIN
+972 Bytes
...contributions/cgcnn_model/AI-SinglePropertyPrediction-Tc_supercon-dft_3d-test-mae.csv.zip
Binary file not shown.