-
Notifications
You must be signed in to change notification settings - Fork 0
Learning about the KB
-
“Short Course: Phylogenetic Comparative Analysis of Integrated Anatomical Traits:”. This tutorial covers using the KB user interface, RPhenoscaPe and PARAMO to access phenotypes and associated semantic data from the KB. The materials are available in RMarkdown files that can be run interactively in the R environment, or the rendered versions can be viewed for RPhenoscape here and PARAMO here.
-
Vignettes in the rphenoscate package demonstrate how to use the R packages rphenoscaTe and rphenoscaPe to perform semantic-aware evolutionary analyses of morphological data.
-
Phenex youtube tutorial. Short demo videos on using Phenex to annotate phylogenetic matrices with ontology terms using the Entity–Quality syntax for describing phenotypes.
- Phenoscape KB web user interface - browse phenotype annotations in the KB by anatomy, quality, taxonomy, and publication.
- Phenoscape API. Phenoscape's publicly accessible web service APIs with service documentation and automatically generated code examples.
- RPhenoscape - R package that facilitates interfacing with the Phenoscape KB for searching ontology terms, querying data matrices, obtaining dependency matrices, and computing semantic similarity metrics.
- rphenoscate - R package that integrates anatomy ontologies in evolutionary models, including functions for assessing anatomical dependencies and semantic similarity metrics for phenotypes, and generating phylogenetic matrices based on mutual exclusivity of semantic phenotypes.
- OntoTrace - tool used to query the KB for a synthetic character matrix of asserted and inferred presence/absence characters for anatomical structures. OntoTrace is also accessible by using RPhenoscape.
- PARAMO - R package for reconstructing organismal ancestral anatomies by modeling anatomical dependencies and using ontology-informed amalgamation of stochastic maps to reconstruct phenotypic evolution at different levels of anatomical hierarchy.
-
The Phenoscape KB contains ontology-annotated phenotypic data for vertebrates and their skeletal features curated from published phylogenetic studies. Examples of data content along the axes of taxonomy and anatomy are given below.
-
Top: data query results for a taxonomic group, catfishes (Order Siluriformes)
-
Bottom: data query results for a trait, parts of the neurocranium
-
-
Most API methods return data in JSON format. The structure of the returned data for each of the services is documented in the REST API documentation (see above). Typically, the identifier for a data item is returned as an IRI in the value of the
@idproperty. Services that return lists of results place the list inside a top-levelresultsproperty. Services that support paging of results will return the total items available (instead of returning results) when thetotal=trueparameter is included. These results will return a single integer as the value of thetotalproperty.Several services support returning TSV via content-negotiation (documented in the response section of the API documentation). TSV can be obtained by requesting the
text/plaincontent type. If JSON is desired, anapplication/jsoncontent type should always be requested in the Accept header, since additional return formats may be added in the future. -
API methods that return a data matrix return data in NeXML format. The following papers may be useful in this regard:
-
Vos RA, Balhoff JP, Caravas JA, Holder MT, Lapp H, Maddison WP, et al. NeXML: rich, extensible, and verifiable representation of comparative data and metadata. Syst Biol. 2012;61: 675–689. doi:10.1093/sysbio/sys025
A description of the NeXML format as an exchange standard for comparative evolutionary analysis.
-
Boettiger C, Chamberlain S, Vos R, Lapp H. RNeXML: a package for reading and writing richly annotated phylogenetic, character and trait data in R. Methods Ecol Evol. 2016;7: 352–357. doi:10.1111/2041-210X.12469
A description of how the R package RNeXML does the heavy lifting of parsing NeXML in R to make it easy to consume for R users. Unfortunately there currently aren't comparable packages/libraries in other languages.
-
-
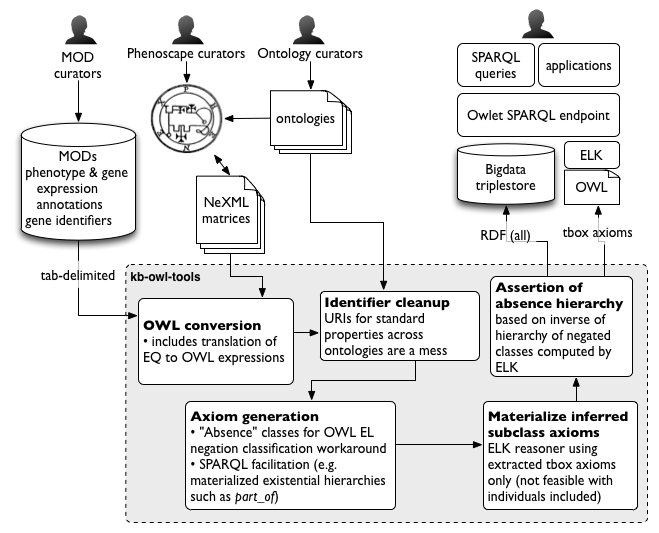
Schematic diagram of the data flow into and out of the KB (Figure 1 in Balhoff et al, 2016). The KB build process is also described on the Phenoscape Wiki.
-
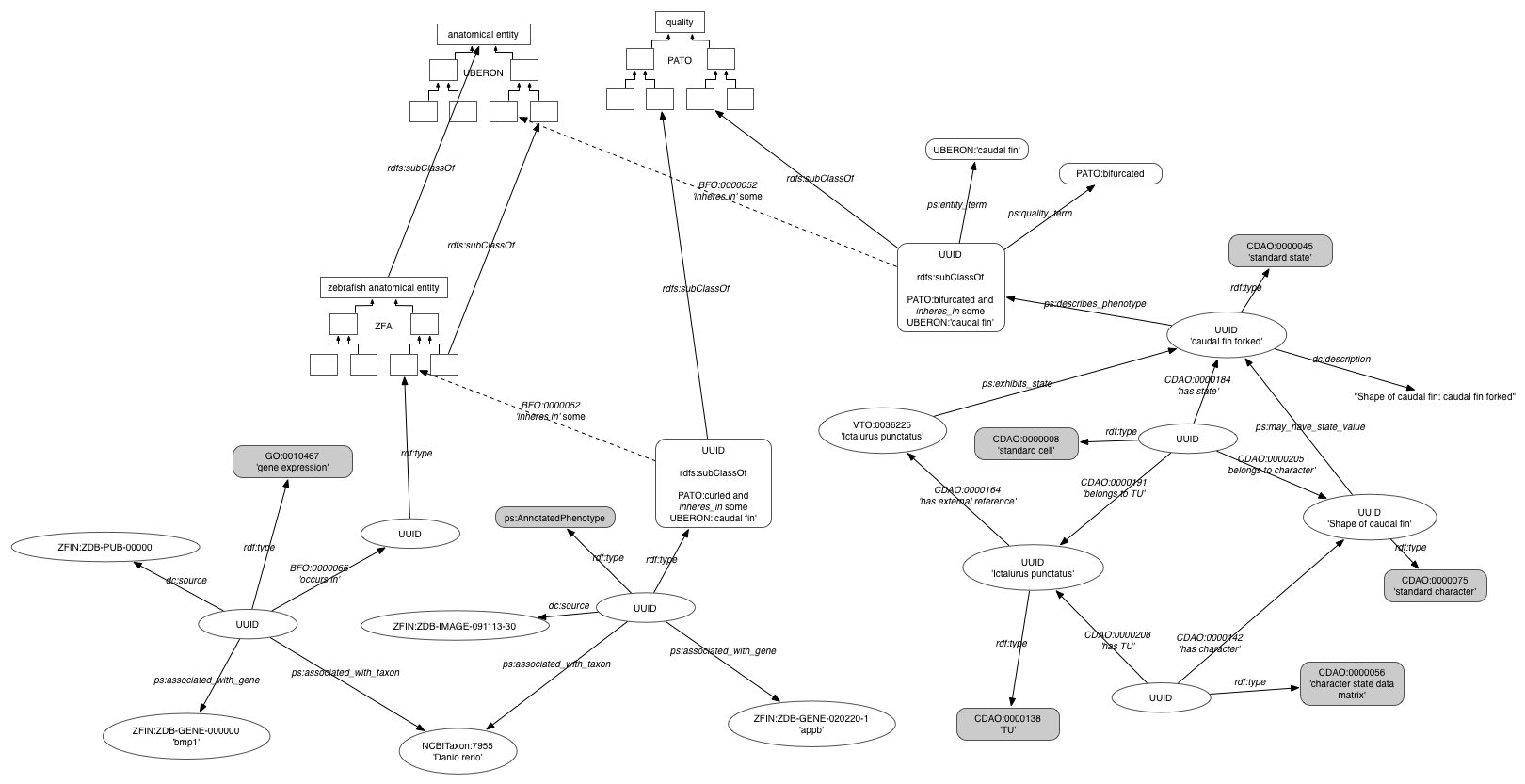
Schematic of the triple model in which data are represented in the KB (high-res version):
-
Porto, D.S., Dahdul, W.M., Lapp, H., Balhoff, J.P., Vision, T.J., Mabee, P.M., and Uyeda, J. (2022) Assessing bayesian phylogenetic information content of morphological data using knowledge from anatomy ontologies. Systematic Biology. doi:10.1093/sysbio/syac022
Paper that describes evaluating phylogenetic information across characters for correspondence to the hierarchical structure and dependencies of anatomy, using anatomical dependencies and semantic similarity distance matrices obtained from Phenoscape KB.
-
Fernando, P.C., Mabee, P.M., Zeng, E. (2021) Gene network module changes associated with the vertebrate fin to limb transition. bioRxiv 2021.01.28.428646; doi:10.1101/2021.01.28.428646
Paper that describes a network-based computational method for identifying gene module changes at key vertebrate evolutionary transitions.
-
Tarasov, S. (2021) New phylogenetic Markov models for inapplicable morphological characters bioRxiv 2021.04.26.441495 doi:10.1101/2021.04.26.441495
Paper that describes the models used in PARAMO and rphenoscate for analysis of anatomically dependent morphological characters. For additional descriptions of the models, see the corHMM vignette.
-
Tarasov, S., Mikó, I., Yoder, M.J., Uyeda, J.C. (2019) PARAMO: A pipeline for reconstructing ancestral anatomies using ontologies and stochastic mapping. Insect Systematics and Diversity, Volume 3, Issue 6, doi:10.1093/isd/ixz009
Paper that demonstrates the use of anatomical dependencies such as those available through Phenoscape KB to reconstruct the evolution of anatomical entities at different hierarchical levels.
-
Jackson, L.M., Fernando, P.C., Hanscom, J.S., Balhoff, J.P., Mabee, P.M. (2018) Automated integration of trees and traits: a case study using paired fin loss across teleost fishes. Systematic Biology, 67(4):559–575, doi:10.1093/sysbio/syx098
Example of applying OntoTrace to generate a synthetic presence-absence matrix for a large scale macroevolutionary study.
-
Balhoff JP, Phenoscape Project Team. The Phenoscape Knowledgebase: tools and APIs for computing across phenotypes from evolutionary diversity and model organisms. bioRxiv. 2016. p. 071951. doi:10.1101/071951
A short read (2 pages) that gives an overview of the data sources and ontologies going into building the KB, the tools and steps involved in building, and the web-service interfaces.
-
Dececchi TA, Balhoff JP, Lapp H, Mabee PM. Toward synthesizing our knowledge of morphology: using ontologies and machine reasoning to extract presence/absence evolutionary phenotypes across studies. Syst Biol. 2015;64: 936–952. doi:10.1093/sysbio/syv031
Paper describing how the KB uses machine reasoning to synthesize presence/absence characters that are implied but not expressly stated by published phenotype descriptions.
-
Manda P, Balhoff JP, Lapp H, Mabee P, Vision TJ. Using the phenoscape knowledgebase to relate genetic perturbations to phenotypic evolution. Genesis. 2015;53: 561–571. doi:10.1002/dvg.22878
Paper describing how algorithms for calculating semantic similarity and significance allow obtaining taxa with evolutionary phenotype transitions semantically similar to the phenotypes of a gene when mutated or knocked out.
Sponsored by Phenoscape. This event is supported by NSF grant DBI-1661456. Phenoscape is also supported by NSF grants DBI-1661529, DBI-1661516, and DBI-1661356. It was originally incubated and supported by the National Evolutionary Synthesis Center (NESCent), NSF EF-0905606.