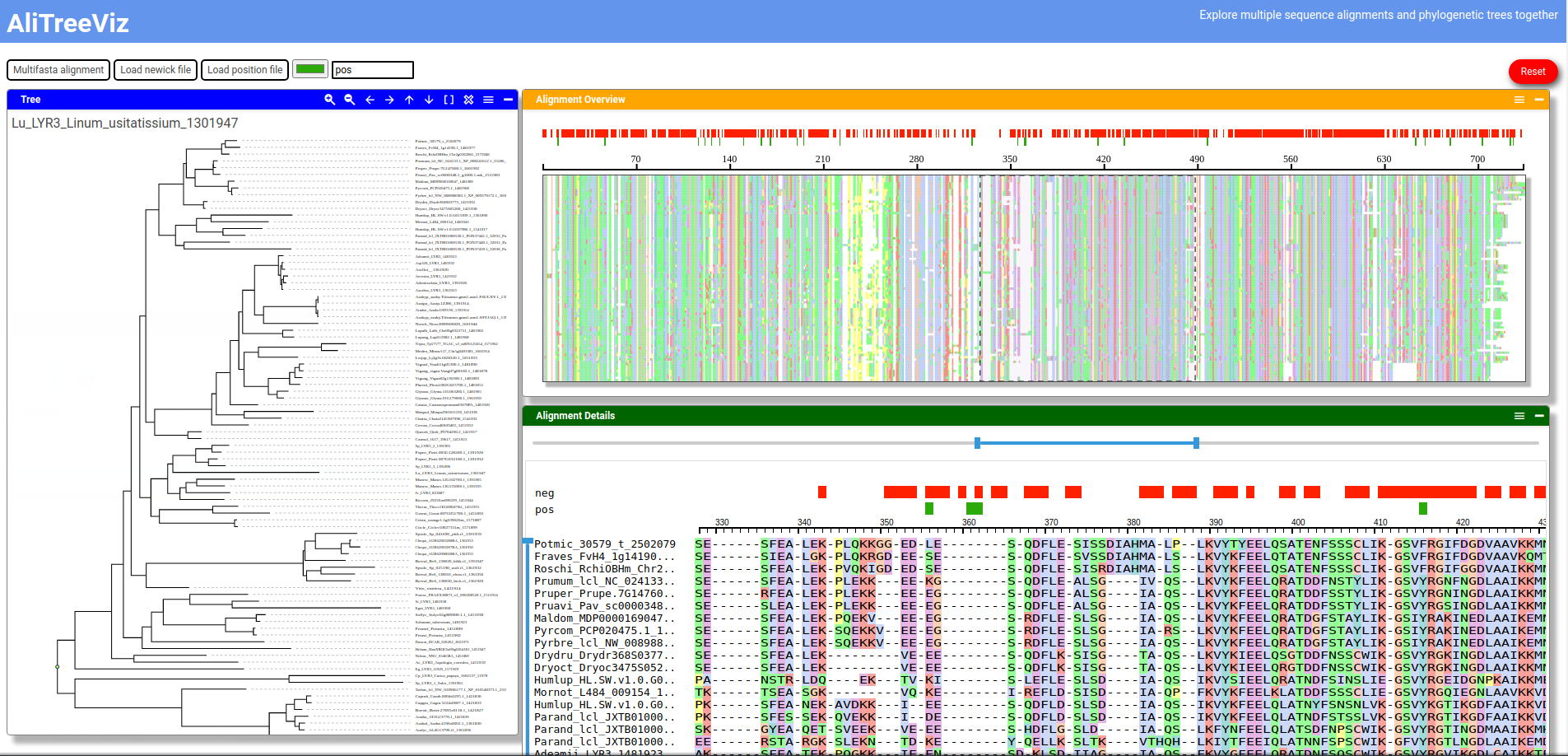
Be careful, this a very preliminary version that could contain bugs.
Visualisation of multiple sequence alignments and phylogenetic trees.
- Does not need any server
- Display phylogenetic tree (newick format)
- Display multiple sequence alignment (MSA) (multifasta format)
- Display MSA overview
- Select sequences in the MSA from the phylogenetic tree (the names of the sequences must correspond)
- Display position tracks above the MSA overview and detailed view (text file with one position per row)
Here : https://lipm-browsers.toulouse.inrae.fr/alitreeviz/
You need npm installed on your computer
git clone https://github.com/lipme/alitreeviz.git
npm install
npm run serve
Now you can use alitreeviz directly since it does not require any server.
npm run build
npm install --save alitreeviz
<template>
<alitreeviz />
</template>
<script>
import Alitreeviz from './views/Alitreeviz.vue'
import 'alitreeviz/dist/alitreeviz.css'
export default {
name: 'App',
components: {
Alitreeviz
}
}
</script>
Note: For the moment, I don't know why, margins might be added so that the component becomes centered in the page...
npm install
npm run serve
npm run build