DeepLabCut is a toolbox for markerless pose estimation of animals performing various tasks, like trail tracking, reaching in mice and various Drosophila behaviors during egg-laying (see Mathis et al. for details). There is, however, nothing specific that makes the toolbox only applicable to these tasks and/or species. The toolbox has also already been successfully applied (by us and others) to rats, humans, various fish species, bacteria, leeches, various robots, cheetahs, mouse whiskers and race horses. This work utilizes the feature detectors (ResNets + readout layers) of one of the state-of-the-art algorithms for human pose estimation by Insafutdinov et al., called DeeperCut, which inspired the name for our toolbox (see references below).
VERSION 2.0: This is the Python package of DeepLabCut. This package includes graphical user interfaces to label your data, and take you from data set creation to automatic behavioral analysis. It also introduces an active learning framework to efficiently use DeepLabCut on large experimental projects.
VERSION 1.0: The initial, Nature Neuroscience version of DeepLabCut can be found in the history of git, or here: https://github.com/AlexEMG/DeepLabCut/releases/tag/1.11
Please check out www.mousemotorlab.org/deeplabcut for more video demonstrations of automated tracking. Above: courtesy of the Murthy (mouse), Leventhal (rat), and Axel (fly) labs!
- How to install DeeplabCut
- An overview of the pipeline and workflow for project management
- We provide several Jupyter Notebooks: one that walks you through a demo dataset to test your installation, and another Notebook to run DeepLabCut from the begining on your own data. We also show you how to use the code in Docker, and on Google Colab. Please also read the user-guide preprint!
- Nov 2018: We posted a detailed guide for DeepLabCut 2.0 on BioRxiv. It also contains a case study for 3D pose estimation in cheetahs.
- Nov 2018: Various (post-hoc) analysis scripts contributed by users (and us) will be gathered at DLCutils. Feel free to contribute! In particular, there is a script guiding you through importing a project into the new data format for DLC 2.0
- Oct 2018: new pre-print on the speed video-compression and robustness of DeepLabCut on BioRxiv
- Sept 2018: Nature Lab Animal covers DeepLabCut: Behavior tracking cuts deep
- Kunlin Wei & Konrad Kording write a very nice News & Views on our paper: Behavioral Tracking Gets Real
- August 2018: Our preprint appeared in Nature Neuroscience
- August 2018: NVIDIA AI Developer News: AI Enables Markerless Animal Tracking
- July 2018: Ed Yong covered DeepLabCut and interviewed several users for the Atlantic.
- April 2018: first DeepLabCut preprint on arXiv.org
-
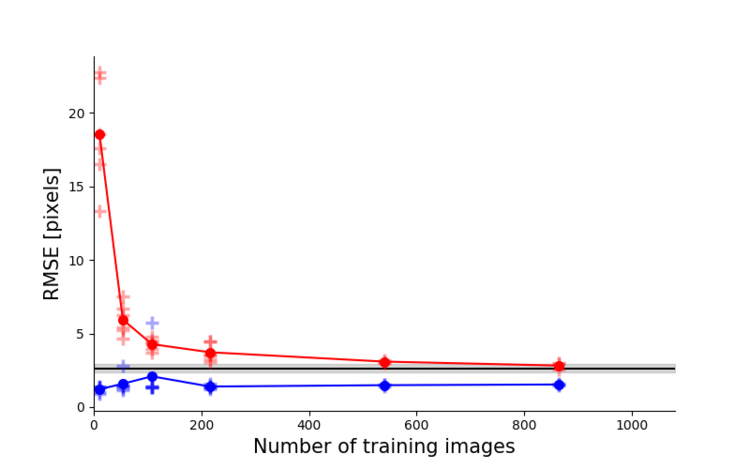
Top left: Due to transfer learning it requires little training data for multiple, challenging behaviors (see Mathis et al. for details).
-
Top Right: Video anlysis is fast (see Mathis/Warren for details)
-
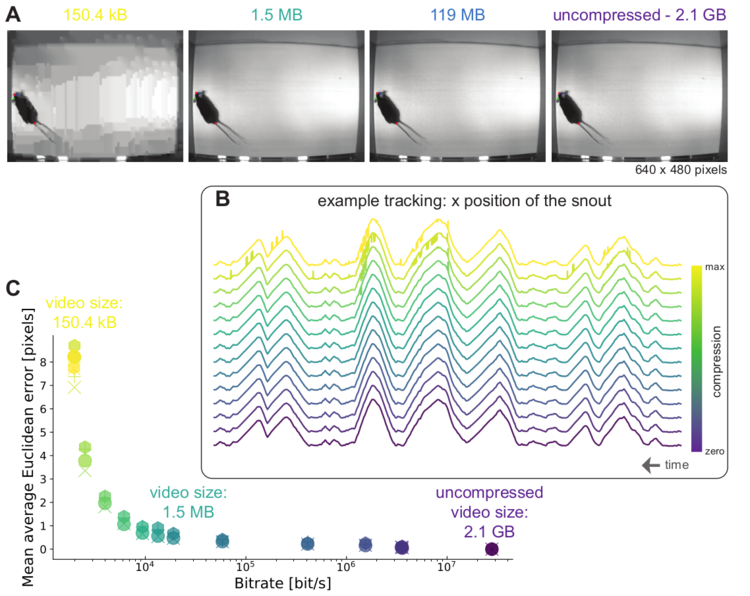
Mid Left: The feature detectors are robust to video compression (see Mathis/Warren for details)
-
Mid Right: It allows 3D pose estimation with a single network and camera (see Mathis/Warren for details)
-
Bottom: It allows 3D pose estimation with a single network trained on data from multple cameras together with standard triangulation methods (see Nath* and Mathis* et al. for details)
Alexander Mathis, Tanmay Nath, Mackenzie Mathis. The feature detector code is based on Eldar Insafutdinov's TensorFlow implementation of DeeperCut. DeepLabCut is an open-source tool and has benefited from suggestions and edits by many individuals including Richard Warren, Ronny Eichler, Jonas Rauber, Hao Wu, Federico Claudi, Gary Kane, Taiga Abe, and Jonny Saunders as well as the contributors. In particular, the authors thank Ronny Eichler for input on the modularized version.
This is an actively developed package and we welcome community development and involvement.
For help and questions that don't fit a GitHub code issue, we ask you to post them here: https://forum.image.sc/
If you would like to join the code development community, please drop us a note to be invited by emailing: [email protected]
Please check out the following references for more details:
@article{Mathisetal2018,
title={DeepLabCut: markerless pose estimation of user-defined body parts with deep learning},
author = {Alexander Mathis and Pranav Mamidanna and Kevin M. Cury and Taiga Abe and Venkatesh N. Murthy and Mackenzie W. Mathis and Matthias Bethge},
journal={Nature Neuroscience},
year={2018},
url={https://www.nature.com/articles/s41593-018-0209-y}
}
@article{insafutdinov2016eccv,
title = {DeeperCut: A Deeper, Stronger, and Faster Multi-Person Pose Estimation Model},
author = {Eldar Insafutdinov and Leonid Pishchulin and Bjoern Andres and Mykhaylo Andriluka and Bernt Schiele},
booktitle = {ECCV'16},
url = {http://arxiv.org/abs/1605.03170}
}
Our open-access pre-prints:
@article{mathis2018markerless,
title={Markerless tracking of user-defined features with deep learning},
author={Mathis, Alexander and Mamidanna, Pranav and Abe, Taiga and Cury, Kevin M and Murthy, Venkatesh N and Mathis, Mackenzie W and Bethge, Matthias},
journal={arXiv preprint arXiv:1804.03142},
year={2018}
}
@article {NathMathis2018,
author = {Nath*, Tanmay and Mathis*, Alexander and Chen, An Chi and Patel, Amir and Bethge, Matthias and Mathis, Mackenzie W},
title = {Using DeepLabCut for 3D markerless pose estimation across species and behaviors},
year = {2018},
doi = {10.1101/476531},
publisher = {Cold Spring Harbor Laboratory},
URL = {https://www.biorxiv.org/content/early/2018/11/24/476531},
eprint = {https://www.biorxiv.org/content/early/2018/11/24/476531.full.pdf},
journal = {bioRxiv}
}
@article {MathisWarren2018speed,
author = {Mathis, Alexander and Warren, Richard A.},
title = {On the inference speed and video-compression robustness of DeepLabCut},
year = {2018},
doi = {10.1101/457242},
publisher = {Cold Spring Harbor Laboratory},
URL = {https://www.biorxiv.org/content/early/2018/10/30/457242},
eprint = {https://www.biorxiv.org/content/early/2018/10/30/457242.full.pdf},
journal = {bioRxiv}
}
This project is licensed under the GNU Lesser General Public License v3.0. Note that the software is provided "as is", without warranty of any kind, express or implied. If you use this code, please cite us!.