This repository contains my implementation of the "full-volume" model from the paper:
End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography.
Ardila, D., Kiraly, A.P., Bharadwaj, S. et al. Nat Med 25, 954–961 (2019).
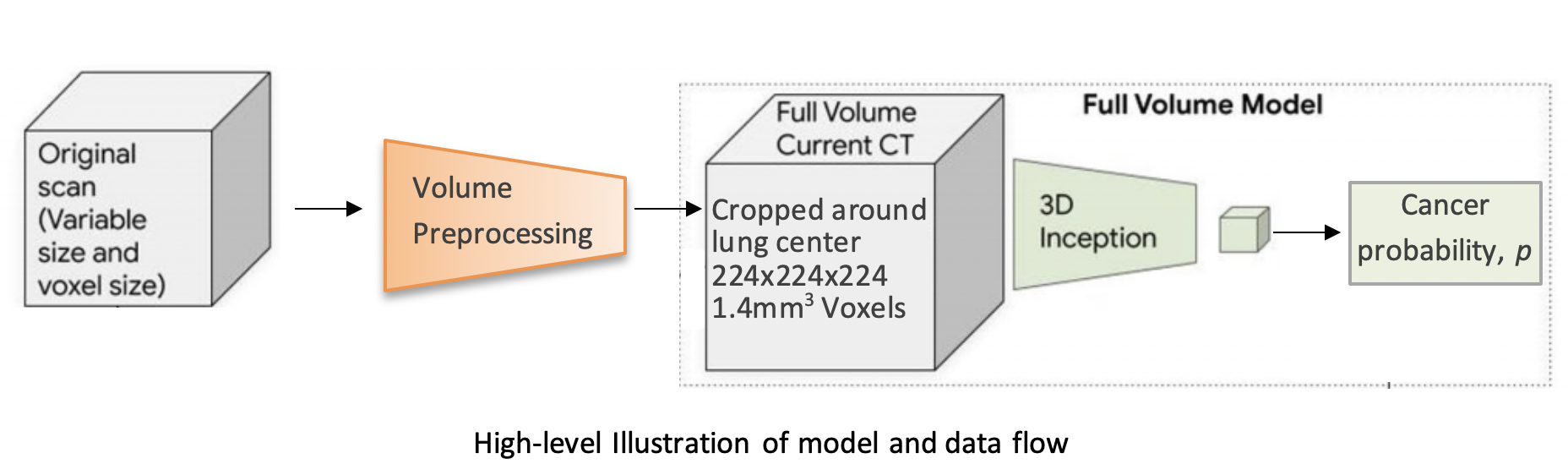
The model uses a three-dimensional (3D) CNN to perform end-to-end analysis of whole-CT volumes, using LDCT volumes with pathology-confirmed cancer as training data. The CNN architecture is an Inflated 3D ConvNet (I3D) (Carreira and Zisserman).
For more detailed information, see this report on arXiv.
We use the NLST dataset which contains chest LDCT volumes with pathology-confirmed cancer evaluations. For description and access to the dataset refer to the NCI website.
pip install lungsimport lungs
lungs.predict('path/to/data')From command line (with preprocessed data):
python main.py --preprocessed --input path/to/preprocessed/dataThe inputs to the training procedure are training and validation .list files containing coupled data - a volume path and its label in each row.
These .list files need to be generated beforehand using preprocess.py, as described in the next section.
import lungs
# train with default hyperparameters
lungs.train(train='path/to/train.list', val='path/to/val.list')The main.py module contains training (fine-tuning) and inference procedures.
The inputs are preprocessed CT volumes, as produced by preprocess.py.
For usage example, refer to the arguments' description and default values in the bottom of main.py.
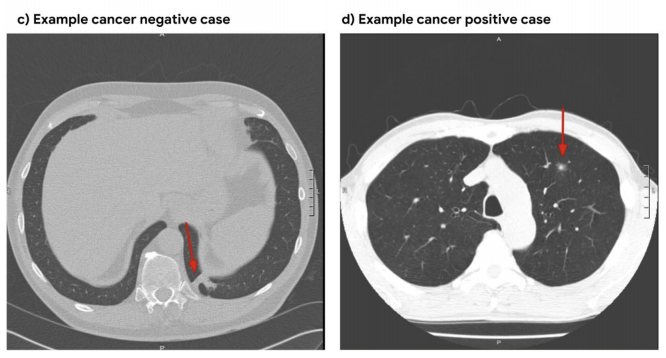
Each CT volume in NLST is a directory of DICOM files (each .dcm file is one slice of the CT).
The preprocess.py module accepts a directory path/to/data containing multiple such directories (volumes).
It performs several preprocessing steps, and writes each preprocessed volume as an .npz file in path/to/data_preprocssed.
These steps are based on this tutorial, and include:
- Resampling to a 1.5mm voxel size (slow)
- Coarse lung segmentation – used to compute lung center for alignment and reduction of problem space
To save storage space, the following preprocessing steps are performed online (during training/inference):
- Windowing – clip pixel values to focus on lung volume
- RGB normalization
Example usage:
from lungs import preprocess
# Step 1: Preprocess all volumes, will save them to '/path/to/dataset_preprocessed'
preprocess.preprocess_all('/path/to/dataset')Once the preprocessed data is ready, the next step is to split it randomly into train/val sets,
and save each set as a .list file of paths/labels, required for the training procedure.
Example usage:
from lungs import preprocess
# Step 2: Split preprocessed data into train/val coupled `.list` files
preprocess.split(positives='/path/to/dataset_preprocessed/positives',
negatives='/path/to/dataset_preprocessed/negatives',
lists_dir='/path/to/write/lists',
split_ratio=0.7)By default, if the ckpt argument is not given, the model is initialized using our best fine-tuned checkpoint.
Due to limited storage and compute time, our checkpoint was trained on a small subset of NLST containing 1,045 volumes (34% positive).
Note that in order to obtain a general purpose prediction model, one would have to train on the full NLST dataset. Steps include:
- Gaining access to the NLST dataset
- Downloading1 ~6TB of positive and negative volumes (requires storage and a few days for downloading)
- Preprocessing (see Data Preprocessing section above)
- Training (requires a capable GPU)
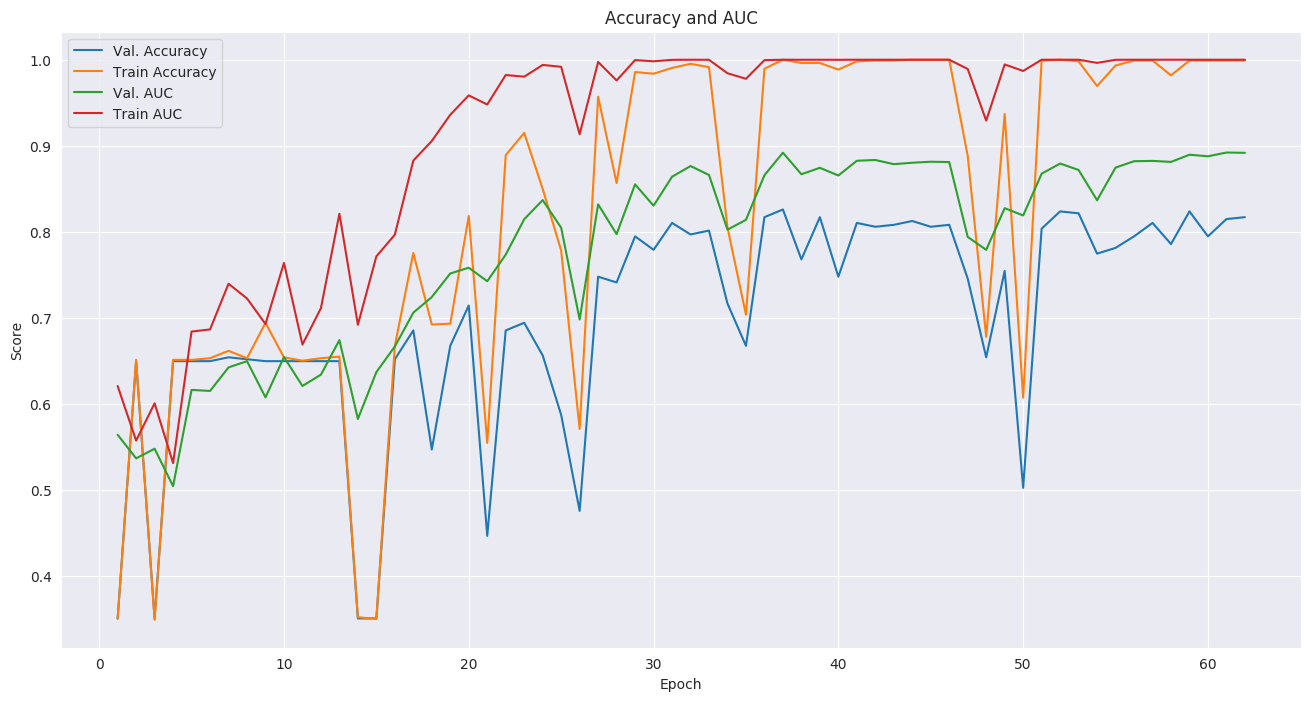
Even though we used a small subset of NLST, we still achieved a state-of-the-art AUC score of 0.892 on a validation set of 448 volumes. This is comparable to the original paper's AUC for the full-volume model (see the paper's supplementary material), trained on 47,974 volumes (1.34% positive).
To train this model we first initialized by bootstrapping the filters from the ImageNet pre-trained 2D Inception-v1 model into 3D, as described in the I3D paper. It was then fine-tuned on the preprocessed CT volumes to predict cancer within 1 year (binary classification). Each of these volumes was a large region cropped around the center of the bounding box, as determined by lung segmentation in the preprocessing step.
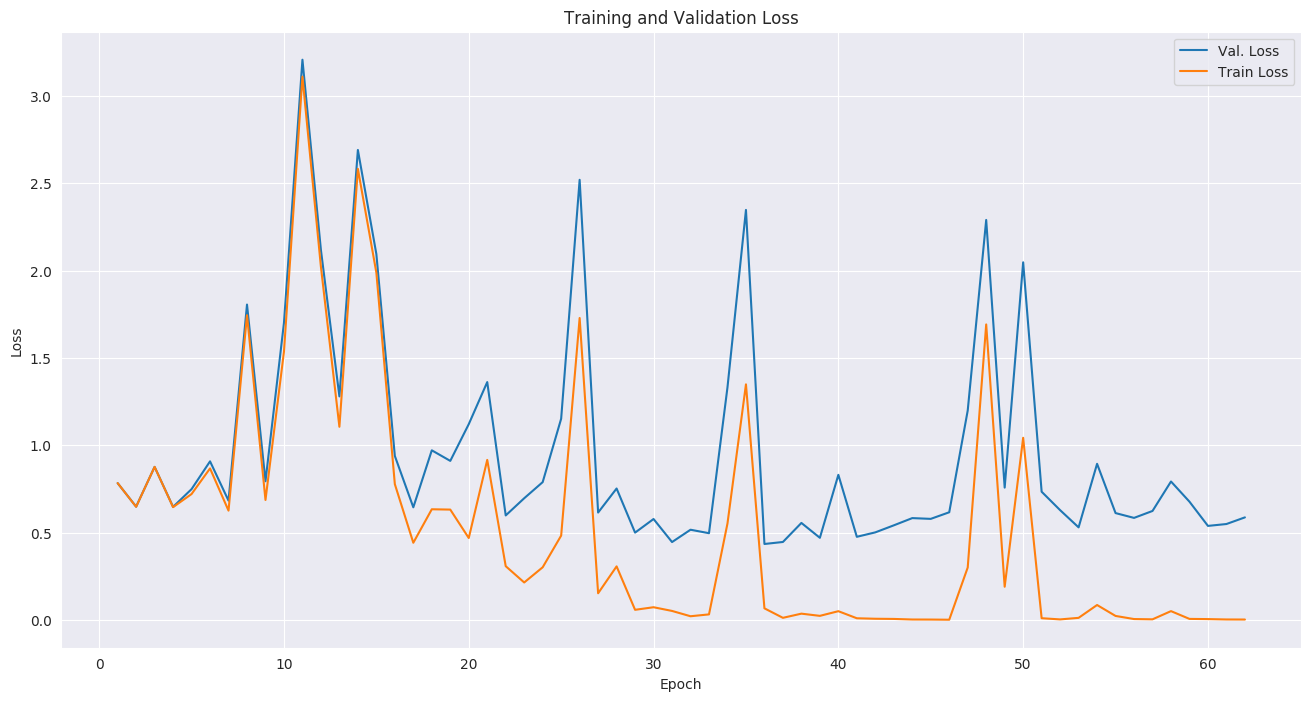
For the training setup, we set the dropout keep_prob to 0.7, and trained in mini-batches of size of 2 (due to limited GPU memory). We used tf.train.AdamOptimizer with a small learning rate of 5e-5, (due to the small batch size) and stopped the training before overfitting started around epoch 37.
The focal loss function from the paper is provided in the code, but we did not experience improved results using it, compared to cross-entropy loss which was used instead. The likely reason is that our dataset was more balanced than the original paper's.
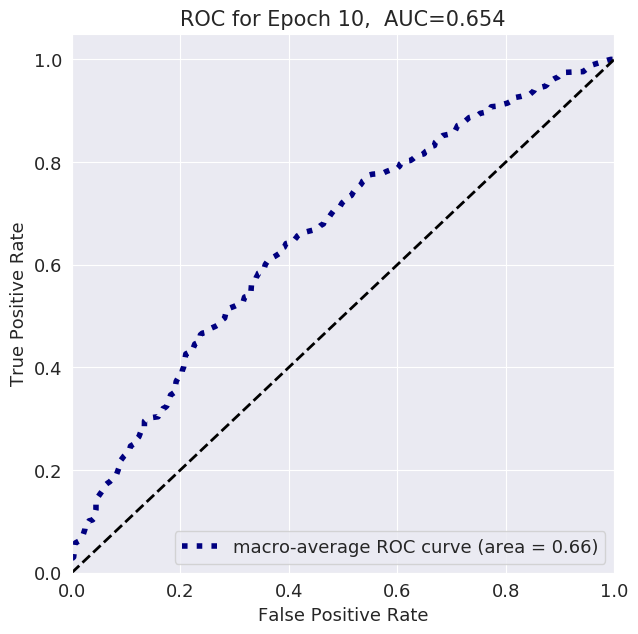
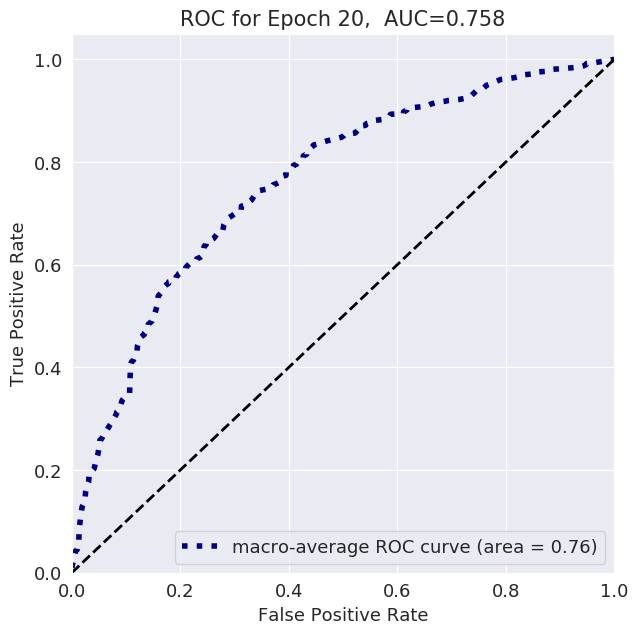
The follwoing plots show loss, AUC, and accuracy progression during training, along with ROC curves for selected epochs:
@software{korat-lung-cancer-pred,
author = {Daniel Korat},
title = {{3D Neural Network for Lung Cancer Risk Prediction on CT Volumes}},
month = jul,
year = 2020,
publisher = {Zenodo},
version = {v1.0},
doi = {10.5281/zenodo.3950478},
url = {https://doi.org/10.5281/zenodo.3950478}
}The author thanks the National Cancer Institute for access to NCI's data collected by the National Screening Trial (NLST). The statements contained herein are solely those of the author and do not represent or imply concurrence or endorsement by NCI.
1 Downloading volumes is done by querying the TCIA website (instruction on NCI website). We used the following query filters:
LungCancerDiagnosis.conflc == "Confirmed.."(positive) or"Confirmed Not..."(negative)SCTImageInfo.numberimages >= 130(minimum number of slices)SCTImageInfo.reconthickness < 5.0(maximum slice thickness)ScreeningResults.study_yr = x(study year of volume, a number between 0 and 7)LungCancerDiagnosis.cancyr = xorx + 1(for positives: study year of patient's cancer diagnosis is equal tostudy_yror 1 year later)
GitHub Pages
arXiv Report
Presentation Video
Presentation PDF
DOI: https://doi.org/10.5281/zenodo.3950478