Here are examples for running Python code on CSC's Puhti supercomputer as different job styles:
- Interactive - for developing your scripts, usually with limited test data.
- Via batch jobs - for computationally more demanding analysis:
- Serial
- Parellel:
- With different Python libraries:
multiprocessing,joblibanddask. - With external tools like
GNU parelleland array jobs.
- With different Python libraries:
If unsure, start with Dask, it is one of the newest, most versatile and easy to use. But Dask has many options and alternatives, multiprocessing might be easier to learn at first and is included in all Python installations by default. multiprocessing and joblib are suitable for maximum of one node (max 40 cores in Puhti). Dask can run also multi-node jobs.
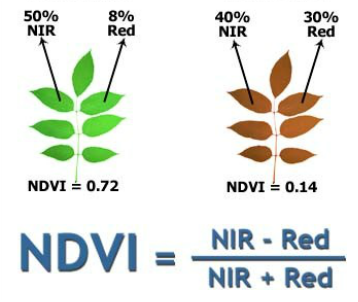
The example calculates NDVI (Normalized Difference Vegetation Index) from Sentinel-2 satellite image's red and near infrared bands. The reading, writing and calculation of NDVI are identical in all examples (with the exception of the Dask - xarray example) and only the method of parallelization changes (the code in the main function).
Basic idea behind the script is to:
- Find red and near infrared channels of Sentinel L2A product from its
SAFEfolder (to simplify, we will call this a Sentinel-2 file from here on) and open thejp2files. ->readImage - Read the data as
numpyarray withrasterio, scale the values to reflectance values and calculate NDVI index. ->calculateNDVI - Save output as GeoTiff with
rasterio. ->saveImage
Files in this example:
- The input ESA Sentinel L2A images are in JPG2000 format and are already stored in Puhti:
/appl/data/geo/sentinel/s2_example_data/L2A. Puhti has only these example images, more Sentinel L2A images are available from CSC Allas. - The example scripts are in subfolders by job type and used parallel library. Each subfolder has 2 files:
- A .py file for defining the tasks to be done.
- A batch job .sh file for submitting the job to Puhti batch job scheduler (SLURM).
If you haven't yet, get the example scripts to Puhti:
Warning
Please note that you will need to adapt the paths mentioned in this material. In most cases it is cscusername and project_20xxxxx that need to be replaced with your CSC username and your projectname.
- Log in to Puhti web interface: https://puhti.csc.fi
- Start a
Login node shell - Create a folder for yourself:
- Switch to your projects scratch directory and further into the students directory:
cd /scratch/project_20xxxxx/students - Create new own folder:
mkdir cscusername - Switch into your own folder:
cd cscusername - Get the exercise material by cloning this repository:
git clone https://github.com/csc-training/geocomputing - Switch to the directory with the example files:
cd geocomputing/python/puhti. - Check that we are in the correct place:
pwdshould show something like/scratch/project_20xxxxx/students/cscusername/geocomputing/python/puhti.
- Switch to your projects scratch directory and further into the students directory:
Within an interactive job it is possible to edit the script in between test-runs; so this suits best when still writing the script. Usually it is best to use some smaller test dataset for this. We can open and look at the Python script from Puhti web interface > Files tab, within Jupyter environment, Visual Studio Code (VSCode) application or the command line. In addition to being able to run the full script within Visual Studio Code, it also possible to run parts of a script step by step.
- Start Visual Studio Code in Puhti web interface.
- Open VSCode start page from front page: Apps -> Visual Studio Code
- Choose settings for VSCode:
- (Reservation:
geocomputing_thu, only during course) - Project:
project_20xxxxx - Partition:
interactive(smallduring course) - Number of CPU cores: 1
- Memory: 2 Gb
- Local disk: 0
- Time: 1:30:00
- Code version and compiler: leave default
- Modules: geoconda
Launch
- (Reservation:
- Wait a moment -> Connect to Visual Studio Code
- VSCode opens up in the browser
- Open folder with the exercise files:
- Click three lines up left -> File -> Open folder ->
/scratch/project_20xxxxx/students/cscusername/geocomputing/python/puhti-> OK
- Click three lines up left -> File -> Open folder ->
- Open 00_interactive/interactive_single_core_example.py. This is a Python script, which calculates NDVI for one input Sentinel-2 "file".
- Check that needed Python libraries are available in Puhti:
- Select all import rows and press
Shift+Enter. Wait a few seconds. The import commands are run in Terminal (which opens automatically on the bottom of the page). If no error messages are visible, the packages are available. Also other parts of the script can be tested in the same manner (select the code and run withShift+Enter).
- Select all import rows and press
- We can also run the full script by following these steps:
- Exit Python console in Terminal: type
exit()in the terminal, if you ran pieces of the code before. - Click the arrow up right above script (Run Python File in Terminal)
- Wait, it takes a few minutes for complete. The printouts will appear in the terminal during the process. Note the time it took.
- Check that there is one new GeoTiff file in your work directory in the Files panel of VSCode.
- Exit Python console in Terminal: type
All computationally heavy analysis should be done via batch jobs. The login node is used only to tell the supercomputer, what it should do. The execution of the code will happen on a compute node -> We go non-interactive. From this point onwards we will have to work from the command line.
-
Open
/scratch/project_20xxxxx/students/cscusername/geocomputing/python/puhti/01_serial/single_core_example.shwith VSCode. -
Check out the changes in the Python file compared to the
00_interactive/interactive_single_core_example.py:- Python script reads one input image file from the argument, which is set inside the batch job file.
Note
Can you find out from the .sh file:
- Where are output and error messages written?
- How many cores are reserved, and for how long a time?
- How much memory?
- Which partition is used?
- Which module is used?
- Does this all sound familiar? Yes, we have given these parameters before, just in a different way.
- Update the project with the number of your project
- Submit the batch job from login node shell (not VSCode terminal or compute node shell):
cd /scratch/project_20xxxxx/students/cscusername/geocomputing/python/puhti/01_serial
sbatch single_core_example.sh
-> sbatch prints out a job ID
Note
Use the job ID to check the state and efficiency of the batch job with seff jobid (replace jobid with the job ID from the print out)
Did you reserve a good amount of memory?
- Once the job is finished, see output in
slurm-jobid.outandslurm-jobid.errwith VSCode for any possible errors and other outputs. - Check that you have a new GeoTiff file in the output folder.
We can also paralellize within Python. In this case Python code takes care of dividing the work to 3 processes running at the same time, one for each input file. Python has several packages for code parallelization, here examples for dask are provided. Here are two examples for single-node (max 40 cores) and multi-node usage.
- Please see Geocomputing course's Dask section for more general introduction.
The examples use delayed functions from Dask to parallelize the workload. Typically, if a workflow contains a for-loop, it can benefit from delayed functions. Besides delayed functions, Dask supports also several othe ways of parallelization, including Dask DataFrames and Dask Arrays. For Dask DataFrames, see our dask-geopandas example and for Dask arrays STAC example with Xarray.
This example uses Dask default scheduler. It can only utilize a single node in supercomputer, so on Puhti the maximum number of CPU cores and workers is 40.
You need to set your project to the batch job file, otherwise this example works out of the box.
-
06_parallel_dask/single_node/dask_singlenode.sh batch job file for
dask.--ntasks=1+--cpus-per-task=3reserves 3 cores - one for each file--mem-per-cpu=2Greserves memory per core
Note
Submit the parallel job to Puhti from login node shell
cd /scratch/project_20xxxxx/students/cscusername/geocomputing/python/puhti/06_parallel_dask/single_node
sbatch dask_singlenode.sh
Note
Check with seff: How much time and resources did you use?
If you want to define more settings, you can also change to Local Distributed scheduler, see the STAC example for details.
This example spreads the workload to several nodes through the Dask-Jobqueue library.
The batch job file launches basically a master job that does not require much resources. It then launches more SLURM jobs that are used for Dask workers, which do the actual computing. The worker jobs are defined inside the multiple_nodes.py file. Note that they have to queue individually for resources so it is good idea to reserve enough time for the master job so it's not cancelled before the workers finish.
With several SLURM jobs it is possible to use several supercomputer nodes that is not possible with other Python parallelization options presented here.
When the worker jobs finish, they will be displayd as CANCELLED on Puhti which is intended, the master job cancels them.
-
06_parallel_dask/multi_node/dask_multinode.sh batch job file for
dask.--ntasks=1+--cpus-per-task=1reserve resources only for the master node
Note
Submit the parallel job to Puhti from login node shell
cd /scratch/project_20xxxxx/students/cscusername/geocomputing/python/puhti/06_parallel_dask/multi_node
sbatch dask_multinode.sh
Note
Check with seff: How much time and resources did you use?
These are just to demonstrate the difference between single core vs. some kind of parallelism. Depending on the issue, some library might be faster or slower. Note also that the performance of your job may depend on some different factors, like disk load or network speed.
| Example | Jobs | CPU cores / job | Time (min) |
|---|---|---|---|
| single core | 1 | 1 | 02:29 |
| multiprocessing | 1 | 3 | 00:52 |
| dask | 1 | 3 | 00:55 |
| GNU parallel | 1 | 3 | 00:58 |
| array job | 3 | 1 | 01:03 |
If you want to start prototyping and testing in a Jupyter Notebook, you can start with an interactive Jupyter session. Choose Jupyter from the Puhti web interface dashboard or the Apps tab in the Puhti web interface. We can find one such prototype file for calculating the NDVI for one file in the materials called interactive.py. It introduces the packages and workflow for this lesson.
-
Settings for Jupyter:
- (Reservation:
geocomputing_thu, only during course) - Project:
project_20xxxxx - Partition:
interactive(smallduring course) - Number of CPU cores: 1
- Memory: 3 Gb
- Local disk: 0
- Time: 1:00:00
- Python: geoconda
- Module version: default
- Jupyter type: Lab
- Working directory:
/scratch/project_20xxxxx/ Launch
- (Reservation:
-
Wait a moment -> Connect to Jupyter
-
Jupyter opens up in the browser
-
Open folder with the exercise files from the left browser panel
students/cscusername/geocomputing/python/puhtiand find the fileinteractive.ipynb. Execute each code cell one after the other withShift+Enter. -
End the session:
- Close the web tab
- In Active sessions view:
Cancel
If you prefer working in the command line, you can also start a compute node shell directly from the tools tab in Puhti web interface. Choose settings for the interactive session:
- Project: project_20xxxxx
- Number of CPU cores: 1
- Memory: 2 Gb
- Local disk: 0
- Time: 1:00:00
Note
You can also start an interactive session from a login node (by starting a login node shell from tools tab in Puhti web interface or by connecting to Puhti via ssh in your own computer's terminal) with:
sinteractive --account project_20xxxxx --cores 1 --time 02:00:00 --mem 4G --tmp 0
This gives you a compute node shell; you can see "where" you are from your terminal prompt [cscusername@puhti-loginXX] -> login node, [cscusername@rXXcXX] (XX being some numbers) -> compute node.
After getting access to the compute node shell, you can load modules and run scripts "like on a normal linux computer":
Note
Remember to change the project name and your CSC user name in the paths below.
module load geoconda
cd /scratch/project_20xxxxx/students/cscusername/geocomputing/python/puhti/00_interactive
python interactive_single_core_example.py
- Wait, it takes a few minutes for complete. The printouts will appear in the terminal during the process. Note the time it took.
- Check that there is one new GeoTiff file in your work directory with
ls -l output - Close the compute node shell tab in your browser and delete the running job from
my interactive sessionsin the Puhti web interface. - Optional, check your results with QGIS
As mentioned, Jupyter is nice for prototyping and testing, however if we want to use this process as part of a larger script, or make it possible to more easily adapt the script to run on other files or calculate other vegetation indices, we need to generalize it and put the code in a Python script. This means for example to put parts of the code into functions, so that they can be reused. You can find one such cleaned up and generalized script with much more comments in 00_interactive/interactive_single_core_example.py.
What if we want to run the same process for not only one, but all Sentinel-2 files within a folder?
-> We could adapt the sbatch script to run the same script with different input one after another in a for loop in the sbatch file, see 01_serial/single_core_example_list.sh (the Python file stays the same as before).
-> Or we adjust the Python script to take in the data folder instead of just one Sentinel-2 file and loop through all files in the main function (see 01_serial/single_core_example_folder.sh and 01_serial/single_core_example_folder.py). You can run it the same way as the single file script, by adapting the project to your project number in the .sh file.
GNU parallel can help parallelizing a script which otherwise is not parallelized. Instead of looping through the three files available within the batch job or the Python script, we let GNU Parallel handle that step. Checkout how it is used in 02_gnu_parallel/gnu_parallel_example.sh with your favorite editor and adapt the project number.
- Changes in the batch job file compared to the
01_serial/single_core_example_list.sh:--cpus-per-taskparameter is used to reserve 3 CPUs for the job- Instead of a for loop, we use
parallelprogram to read in the filenames from the text file and distribute them to the CPUs - Memory and time allocations
To get to know how many
cpus-per-taskwe need you can use for examplels /appl/data/geo/sentinel/s2_example_data/L2A | wc -lin the login node shell to count everything within the data directory before writing the batch job script.
Submit the job to Puhti from login node shell:
Note
Remember to change the project name and your CSC user name in the paths below.
cd /scratch/project_20xxxxx/students/cscusername/geocomputing/python/puhti/02_gnu_parallel
sbatch gnu_parallel_example.sh
Note
Check with seff jobid: How much time and resources did you use?
In the array job example the idea is that the Python script will run one process for every given input file as opposed to running a for loop within the script or one job on multiple CPUs (GNU parallel).
- 03_array/array_job_example.sh array job batch file. Changes compared to
01_serial/single_core_example_list.sh:--arrayparameter is used to tell how many jobs to start. Value 1-3 in this case means that$SLURM_ARRAY_TASK_IDvariable will be from 1 to 3. We can usesedto read the first three lines from ourimage_path_list.txtfile and start a job for each input file.- Output from each job is written to
slurm-jobid_arrayid.outandslurm-jobid_arrayid.errfiles. - Memory and time allocations are per job.
Note
Submit the array job to Puhti from login node shell
Note
Remember to change the project name and your CSC user name in the paths below.
cd /scratch/project_20xxxxx/students/cscusername/geocomputing/python/puhti/03_array
-
04_parallel_multiprocessing/multiprocessing_example.sh batch job file for
multiprocessing.--ntasks=1+--cpus-per-task=3reserves 3 cores - one for each file--mem-per-cpu=2Greserves memory per core
-
04_parallel_multiprocessing/multiprocessing_example.py
- Note how the pool of workers is started and processes divided to workers with
pool.map(). This replaces the for loop that we have seen in01_serial/single_core_example_folder.sh.
- Note how the pool of workers is started and processes divided to workers with
Note
Submit the parallel job to Puhti from login node shell
cd /scratch/project_20xxxxx/students/cscusername/geocomputing/python/puhti/04_parallel_multiprocessing
sbatch multiprocessing_example.sh
Note
Check with seff: How much time and resources did you use?
sbatch array_job_example.sh
Note
Check with seff: How much time and resources did you use?