-
Notifications
You must be signed in to change notification settings - Fork 42
PicoProducer analysis
The modules under PicoProducer/python/analysis are used to process nanoAOD for analysis. Analysis modules
- pre-select events, e.g. those passing some trigger and containing a muon-tau pair;
- reconstruct variables like invariant mass;
- apply corrections, like energy scale, SFs, weights, etc;
-
save variables in branches of a custom tree (ntuple).
The analysis modules are run on nanoAOD with the post-processors,
for example with
picojob.py. The output is a custom analysis ntuple, we refer to as the pico format.
To run to an analysis module with pico.py, you can link the module to a channel shortname in several ways, e.g.
pico.py channel mutau ModuleMuTau
pico.py channel mutau python/analysis/ModuleMuTau.py
and then run it with e.g.
pico.py run -c mutau -y 2016
For more detailed instructions how to run the analysis code with pico.py, see the PicoProducer wiki page.
To run analysis modules with pico.py, the modules should always by in the
PicoProducer/python/analysis/
directory, but you can organize your modules in subdirectories, e.g.
pico.py channel mutau MyAnalysis.ModuleMyMuTau
pico.py channel mutau python/analysis/MyAnalysis/ModuleMyMuTau.py
Furthermore, the module file should have the exact same name as the module class it contains,
e.g. ModuleMuTau.py contains ModuleMuTau.
Please refer to the nanoAOD documentation
for a full list of available variables.
To know how they are defined from miniAOD, you can dig in the CMSSW source code in
cmssw/PhysicsTools/NanoAOD.
To access information of nanoAOD using python, you can subclass Module
from the nanoAOD-tools.
A simple example of a subclass to analyze nanoAOD is given in
ModuleMuTauSimple.py.
The Module class has some pre-defined methods like beginJob and endJob that are called by
PostProcessor,
but the main routine is in analyze, for example:
from PhysicsTools.NanoAODTools.postprocessing.framework.eventloop import Module
class ModuleMuTauSimple(Module):
def analyze(self,event):
muon_idx = [ ]
for imuon in event.nMuon:
if event.Muon_pt[imuon]>20:
muon_idx.append(imuon)
if len(muon_idx)<1:
return False
return True
The analyze method should return True if the event passes your pre-selection,
and you like to keep its information, and False otherwise.
Without loss of performance, you can make the latter more readable using
Collection:
from PhysicsTools.NanoAODTools.postprocessing.framework.eventloop import Module
from PhysicsTools.NanoAODTools.postprocessing.framework.datamodel import Collection
class ModuleMuTauSimple(Module):
def analyze(self, event):
muons = [ ]
for muon in Collection(event,'Muon'):
if muon.pt>20:
muons.append(muon)
if len(muons)<1:
return False
return True
Triggers are saved as booleans, e.g.
if not event.HLT_IsoMu24:
return False
To reduce the nanoAOD file size, some identification working points (WPs) are saved in nanoAOD as UChar_t, which is 1 byte (8 bits),
instead of 4 bytes (32 bits) like Int_t. For example, to require the Medium WP of the DeepTau2017v2p1VSjet tau identification,
you see in the documentation
that it corresponds to the fifth bit, i.e. 2**(5-1)=16.
Note that if a tau object passes the Medium WP, it also passes all looser ones.
The actual value of Tau_idDeepTau2017v2p1VSjet will therefore be "cumulative", e.g. 1+2+4+8+16=31 for Medium, and not just 16.
To access them in python, you may need the built-in function ord, e.g.
tau_idx = [ ]
for itau in event.nTau:
if event.Tau_pt[itau]>20 and ord(event.Tau_idDeepTau2017v2p1VSjet[itau])>=16:
tau_idx.append(itau)
If you use Collections, you do not need ord anymore:
taus = [ ]
for tau in Collection(event,'Tau'):
if tau.pt>20 and tau.idDeepTau2017v2p1VSjet>=16:
taus.append(tau)
Beside WPs, status flags like the integer GenPart_statusFlags of generator particles
are also encoded bitwise. If you want to know if some flag like isPrompt (0th bit, 1) or isHardProcess (7th bit, 128) is triggered,
use bitwise operators as
isPrompt = (GenPart_statusFlags[i] & 1)>0 # bit 0: 2^0 = 1<<0 = 1
hardProcess = (GenPart_statusFlags[i] & 128)>0 # bit 7: 2^7 = 1<<7 = 128
isBoth = (GenPart_statusFlags[i] & 129)==129 # both simultaneously: 2^0 + 2^7 = 129
which has values True or False. Here, 128 is computed as a power of 2, 2**7, or a bitwise left shift, 1<<7.
The help function hasbit in PicoProducer/python/analysis/utils.py can be used:
isPrompt = hasbit(GenPart_statusFlags[i],0)
hardProcess = hasbit(GenPart_statusFlags[i],7)
For matching trigger objects, please see the PicoProducer/python/corrections/TrigObjMatcher.py tool.
A simple example to make your custom tree is given in ModuleMuTauSimple.py. Reduced:
import numpy as np
class ModuleMuTauSimple(Module):
def __init__(self,fname,**kwargs):
self.outfile = TFile(fname,'RECREATE')
def beginJob(self):
self.tree = TTree('tree','tree')
self.pt_1 = np.zeros(1,dtype='f') # 32-bit float
self.q_1 = np.zeros(1,dtype='i') # 32-bit integer
self.id_1 = np.zeros(1,dtype='?') # 1 byte boolean
self.tree.Branch('pt_1',self.pt_1,'pt_1/F') # Float_t
self.tree.Branch('q_1', self.q_1, 'q_1/I') # Int_t
self.tree.Branch('id_1',self.id_1,'id_1/O') # Boot_t
def analyze(self, event):
self.pt_1[0] = 20.0
self.q_1[0] = -1
self.id_1[0] = True
self.tree.Fill()
return True
def endJob(self):
self.outfile.Write()
self.outfile.Close()
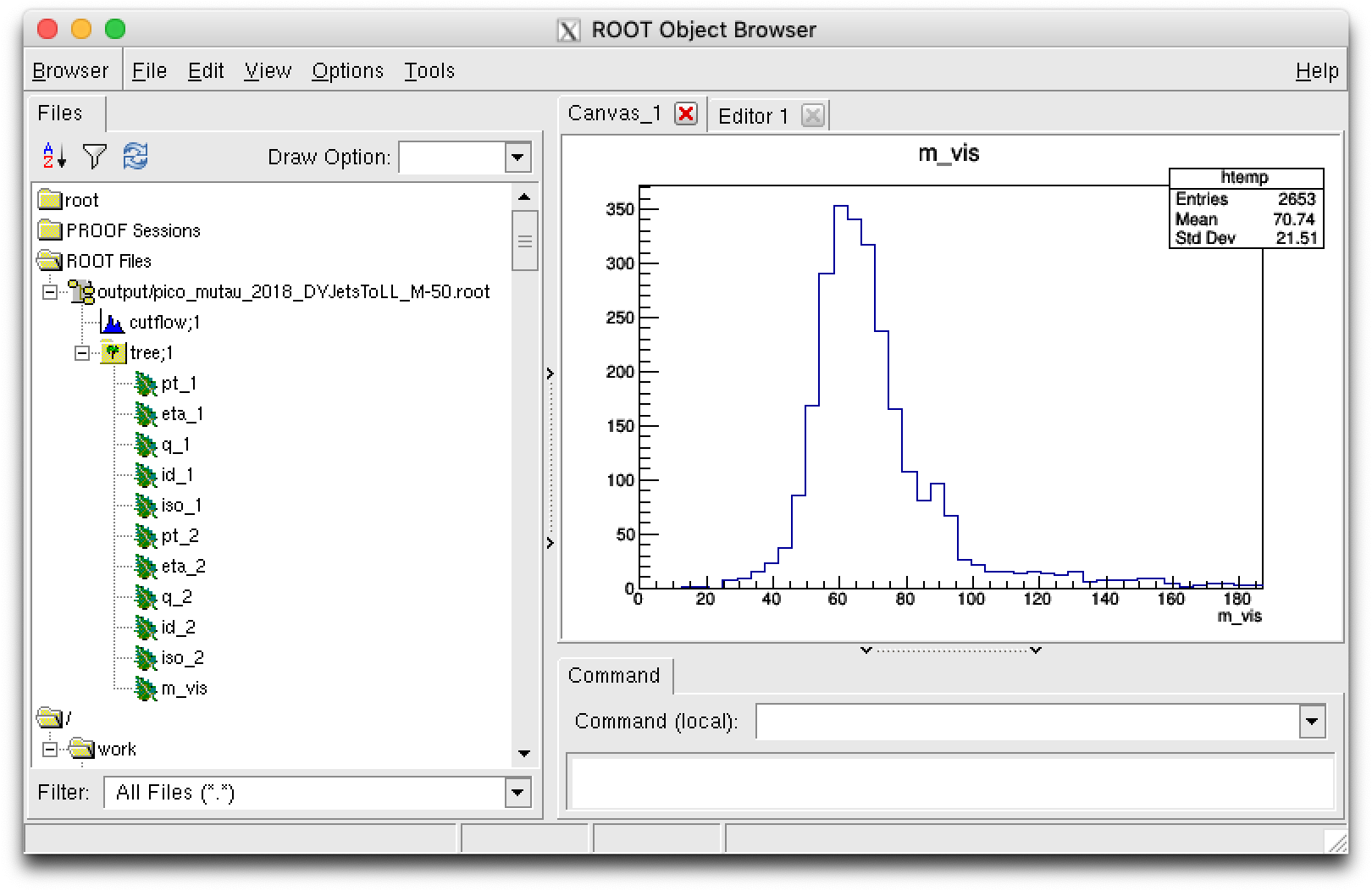
More information on data types for defining the right numpy arrays and branches:
- In
ROOT: https://root.cern.ch/doc/master/classTTree.html#addcolumnoffundamentaltypes - In
numpy: https://numpy.org/devdocs/user/basics.types.html
| ROOT | Branch | numpy |
array |
Data type |
|---|---|---|---|---|
| Bool_t | 'O' |
'?', 'bool', bool
|
8-bit boolean | |
| UChar_t | 'b' |
'b', 'byte', 'int8'
|
'B' |
8-bit unsigned integer |
| Int_t | 'I' |
'i', 'int32'
|
'l' |
32-bit (signed) integer |
| Long64_t | 'L' |
'l', 'int62', long
|
'q' |
64-bit (signed) integer |
| Float_t | 'F' |
'f', 'float32'
|
'f' |
32-bit float |
| Double_t | 'D' |
'd', 'float64', float
|
'd' |
64-bit float |
To make your life easier, you can use separate "tree producer" classes.
For example, TreeProducer can be subclassed as in
TreeProducerMuTau.py.
You then define branches with something like
from TauFW.PicoProducer.analysis.TreeProducer import TreeProducer
class TreeProducerMuTau(TreeProducer):
def __init__(self, filename, module, **kwargs):
super(TreeProducer,self).__init__(filename,module,**kwargs)
self.addBranch('pt_1','f') # float
self.addBranch('q_1', 'i') # integer
self.addBranch('id_1','?') # boolean
In the main analysis module ModuleMuTau.py, you basically do
from TauFW.PicoProducer.analysis.TreeProducerMuTau import TreeProducerMuTau
class ModuleMuTau(Module):
def __init__(self, fname, **kwargs):
self.out = TreeProducerMuTau(fname,self)
def analyze(self, event):
self.out.pt_1[0] = 20.0
self.out.q_1[0] = -1
self.out.id_1[0] = True
self.out.fill()
return True
To add vector branches, use
class TreeProducerMuMu(TreeProducer):
def __init__(self, filename, module, **kwargs):
super(TreeProducer,self).__init__(filename,module,**kwargs)
self.addBranch('pt', 'f',len=2) # vector of fixed length
self.addBranch('njets','i')
self.addBranch('jpt', 'f',len='njets',max=20) # vector of variable length
class ModuleMuMu(Module):
def __init__(self, fname, **kwargs):
self.out = TreeProducerMuMu(fname,self)
def analyze(self, event):
# select two muons
for i, muon in enumerate(muons):
self.out.pt[i] = muon.pt
# prepare jet collection (variable length)
jets = jets[:20] # store maximum 20 jets
self.out.njets[0] = len(jets)
for i, jet in enumerate(jets):
self.out.jpt[i] = jet.pt
self.out.fill()
return True
A full example of usage is shown in the executable
PicoProducer/python/analysis/TestModule.py.
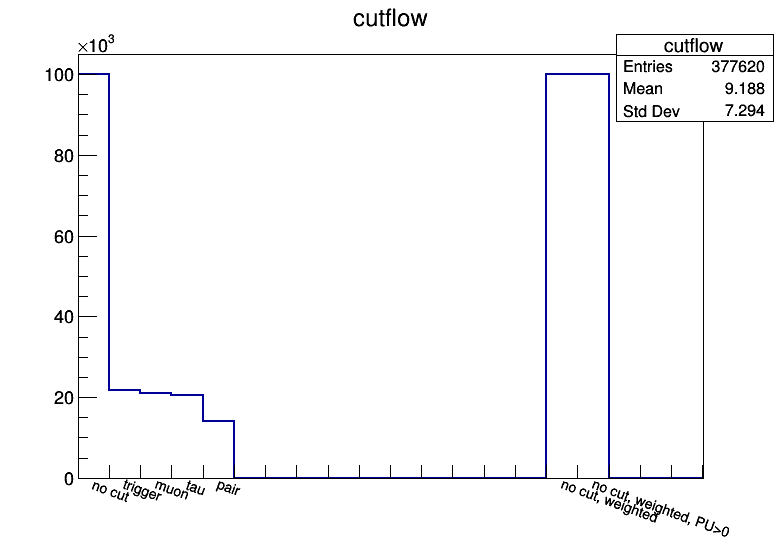
To keep track of the total number of processed events,
and efficiencies of each step in the pre-selection, one should use a cuflow.
This is a simple histogram, binned per integer, that is filled each time a pre-selection is passed.
Again, ModuleMuTauSimple.py provides a straightforward solution.
The TreeProducer class already uses a special Cutflow class,
that can be used as
class ModuleMuTau(Module):
def __init__(self, fname, **kwargs):
self.out = TreeProducer(fname,self)
self.out.cutflow.addcut('none', "no cut" ) # bin 1 (0-1): total number of events
self.out.cutflow.addcut('trig', "trigger" ) # bin 2 (1-2): number of events passing the trigger
self.out.cutflow.addcut('muon', "muon" ) # bin 3 (2-3): number of events with pre-selected muon
self.out.cutflow.addcut('tau', "tau" ) # bin 4 (3-4): number of events with pre-selected tau
self.out.cutflow.addcut('weight',"weight",15) # bin 16 (15-16): total sum of weights (for MC only)
def analyze(self, event):
self.out.cutflow.fill('none')
self.out.cutflow.fill('weight',event.genWeight)
# require trigger, else return False...
self.out.cutflow.fill('trig')
# require muon, else return False...
self.out.cutflow.fill('muon')
# require tau, else return False...
self.out.cutflow.fill('tau')
return True
def endJob(self):
self.out.endJob()
Note that if the generator weight in MC is very different than 1.0 (e.g. for some ttbar samples), it can be useful to save the total sum of weights of all processed events, so it can be used in the denominator of the lumi-cross section normalization, instead of using the total number of MC events.
A set of full analysis modules for the basic study of events with a pair of tau decay candidates (like Z → 𝜏𝜏 events meant for TauPOG studies) are provided. They follow this hierarchy:
-
TreeProducer: general superclass for containing the output file, tree and cutflow-
TreeProducerTauPairprovides common branches in ditau analyses-
TreeProducerMuTaufor the mutau channel -
TreeProducerETaufor the etau channel -
TreeProducerTauTaufor the tautau channel -
TreeProducerMuMufor the mumu channel -
TreeProducerEMufor the emu channel
-
-
-
Module: general superclass for nanoAOD processing-
ModuleTauPairprovides common routines like jet selection or ditau reconstruction (mt,m_vis,dR_ll, etc.)-
ModuleMuTaufor the mutau channel -
ModuleETaufor the etau channel -
ModuleTauTaufor the tautau channel -
ModuleMuMufor the mumu channel -
ModuleEMufor the emu channel
-
-
(Note they are still under construction, and more will be added in the near future.)
Several other modules are stored here:
-
GenDumper.py: To dump some basic generator-level information (PDG ID, pt, status, status flags, ...) of a given sample. -
GenFilterMuTau.py: To study the generator mutau-filter, used for stitching. -
GenMatcher.py: To study the gen-match algorithm for hadronic taus (Tau_genPartFlav). -
PileUp.py: To compile histograms of pileup / number of primary vertices distributions in MC, see these instructions. -
StitchEffs.py: To create 1D & 2D histograms of LHE-level Njets (number of partons) & HT to compute stitching weights forDY*Jets*(HT)&W*Jets*(HT)samples.
Correction tools are found in PicoProducer/python/corrections/ and
corresponding weights, scale factors and more in PicoProducer/data/.