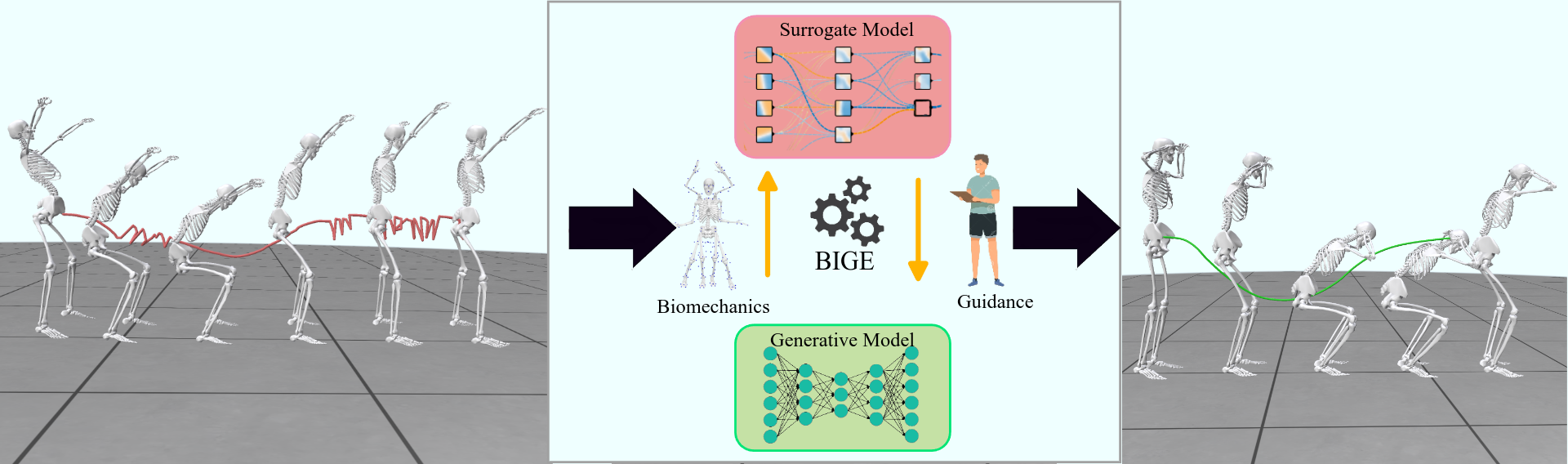
BIGE is a framework for generative models to adhere to clinician-defined constraints. To generate realistic motion, our method uses a biomechanically informed surrogate model to guide the generation process.
Proper movements enhance mobility, coordination, and muscle activation, which are crucial for performance, injury prevention, and overall fitness. However, traditional simulation tools rely on strong modeling assumptions, are difficult to set up and computationally expensive. On the other hand, generative AI approaches provide efficient alternatives to motion generation. But they often lack physiological relevance and do not incorporate biomechanical constraints, limiting their practical applications in sports and exercise science. To address these limitations:
- We propose a novel framework, BIGE, that combines bio-mechanically meaningful scoring metrics with generative modeling.
- BIGE integrates a differentiable surrogate model for muscle activation to reverse optimize the latent space of the generative model.
- Enables the retrieval of physiologically valid motions through targeted search.
- Through extensive experiments on squat exercise data, our framework demonstrates superior performance in generating diverse, physically plausible motions while maintaining high fidelity to clinician-defined objectives compared to existing approaches.
- 1. Visual Results
- 2. Installation
- 3. Quick Start
- 4. Train
- 5. Evaluation
- 6. SMPL Mesh Rendering
- 7. Acknowledgement
- 8. Commands
1. Visual Results (More results can be found in our project page)
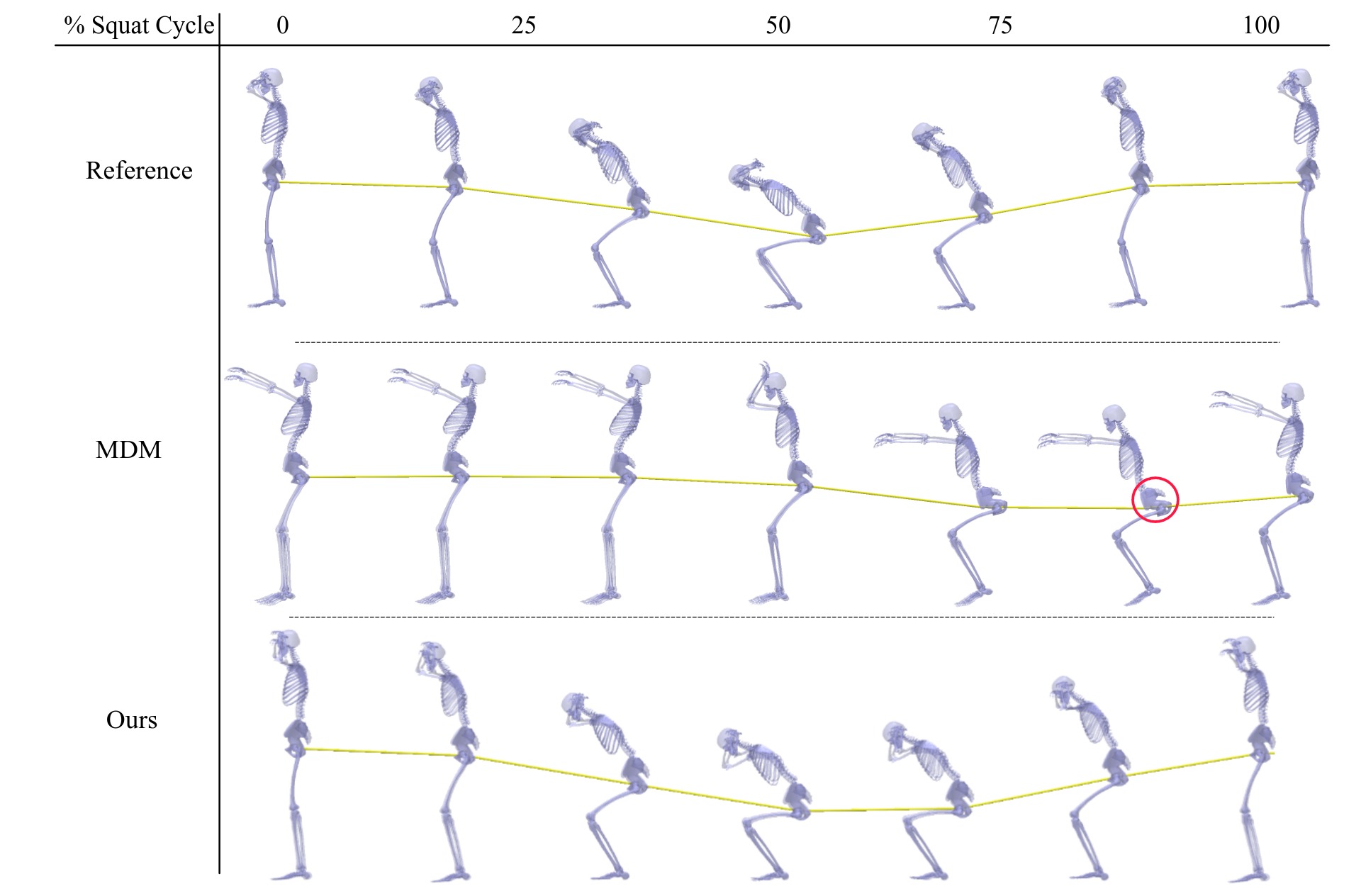
Our guidance strategy leads to a more physiologically accurate squat motion as evidenced by the increased depth of the squat. The generated motion samples are ordered by the peak muscle activation. The red and green lines at 50% squat cycle represent the depth of the squat.
Comparison of generated samples from baselines and BIGE. The yellow curve represents the movement of the hip joint over the entire squat cycle. BIGE generates a more realistic squat motion compared to baselines.
Our model can be learnt in a single GPU V100-32G
conda env create -f environment.yml
conda activate T2M-GPT
ulimit -n 1000000 # Need to run pytorch open multiple files https://stackoverflow.com/questions/71642653/how-to-resolve-the-error-runtimeerror-received-0-items-of-ancdataThe code was tested on Python 3.8 and PyTorch 1.8.1.
More details about the dataset used can be found here [here]
The pretrained model files will be stored in the 'pretrained' folder:
Train the VQVAE model with specified parameters.
python3 train_vq.py --batch-size 256 --lr 2e-4 --total-iter 300000 --lr-scheduler 200000 --nb-code 512 --down-t 2 --depth 3 --dilation-growth-rate 3 --out-dir output --dataname mcs --vq-act relu --quantizer ema_reset --loss-vel 0.5 --recons-loss l1_smooth --exp-name VQVAE9VQVAE Training without DeepSpeed
Train the VQVAE model using DeepSpeed for optimized performance.python3.8 /home/ubuntu/.local/bin/deepspeed train_vq.py --batch-size 256 --lr 2e-4 --total-iter 300000 --lr-scheduler 200000 --nb-code 512 --down-t 2 --depth 3 --dilation-growth-rate 3 --out-dir output --dataname mcs --vq-act relu --quantizer ema_reset --loss-vel 0.5 --recons-loss l1_smooth --exp-name VQVAE9VQVAE Reconstruction
Generate samples using the trained VQVAE model.
python MOT_eval.py --dataname mcs --out-dir output --exp-name VQVAE5_v2 --resume-pth output/VQVAE5_v2/300000.pthRun surrogate training for the model.
python3.8 surrogate_training.pypython LIMO_Surrogate.py --exp-name TestBIGE --vq-name /home/ubuntu/data/T2M-GPT/output/VQVAE14/120000.pth --dataname mcs --seq-len 49 --total-iter 3000 --lr 0.5 --num-runs 3000 --min-samples 20 --subject /data/panini/MCS_DATA/Data/d66330dc-7884-4915-9dbb-0520932294c4 --low 0.35 --high 0.45Run guidance for all subjects
import os
mcs_sessions = ["349e4383-da38-4138-8371-9a5fed63a56a","015b7571-9f0b-4db4-a854-68e57640640d","c613945f-1570-4011-93a4-8c8c6408e2cf","dfda5c67-a512-4ca2-a4b3-6a7e22599732","7562e3c0-dea8-46f8-bc8b-ed9d0f002a77","275561c0-5d50-4675-9df1-733390cd572f","0e10a4e3-a93f-4b4d-9519-d9287d1d74eb","a5e5d4cd-524c-4905-af85-99678e1239c8","dd215900-9827-4ae6-a07d-543b8648b1da","3d1207bf-192b-486a-b509-d11ca90851d7","c28e768f-6e2b-4726-8919-c05b0af61e4a","fb6e8f87-a1cc-48b4-8217-4e8b160602bf","e6b10bbf-4e00-4ac0-aade-68bc1447de3e","d66330dc-7884-4915-9dbb-0520932294c4","0d9e84e9-57a4-4534-aee2-0d0e8d1e7c45","2345d831-6038-412e-84a9-971bc04da597","0a959024-3371-478a-96da-bf17b1da15a9","ef656fe8-27e7-428a-84a9-deb868da053d","c08f1d89-c843-4878-8406-b6f9798a558e","d2020b0e-6d41-4759-87f0-5c158f6ab86a","8dc21218-8338-4fd4-8164-f6f122dc33d9"]
exp_name = "FinalFinalHigh"
for session in mcs_sessions:
os.system(f"python LIMO_Surrogate.py --exp-name {exp_name} --vq-name /data/panini/T2M-GPT/output/VQVAE14/120000.pth --dataname mcs --seq-len 49 --total-iter 3000 --lr 0.5 --num-runs 3000 --min-samples 20 --subject /data/panini/MCS_DATA/Data/{session} --low 0.35 --high 0.45")-
For BIGE
python calculate_wasserstein.py --file_type mot --folder_path /home/ubuntu/data/MCS_DATA/LIMO/FinalFinalHigh/mot_visualization/ -
For motion capture
python wasserstein_mocap.py --file_type mot --folder_path /home/ubuntu/data/MCS_DATA/Data/
-
For baselines
python calculate_wasserstein.py --file_type mot --folder_path /home/ubuntu/data/MCS_DATA/baselines/mdm_baseline/ python calculate_wasserstein.py --file_type mot --folder_path /home/ubuntu/data/MCS_DATA/baselines/t2m_baseline/ python calculate_wasserstein.py --file_type mot --folder_path /home/ubuntu/data/MCS_DATA/LIMO/VQVAE-Generations/mot_visualization/
-
For references
python calculate_guidance.py --file_type mocap --folder_path /home/ubuntu/data/MCS_DATA/Data/
-
For baselines
python calculate_guidance.py --file_type mot --folder_path /home/ubuntu/data/MCS_DATA/baselines/mdm_baseline/ python calculate_guidance.py --file_type mot --folder_path /home/ubuntu/data/MCS_DATA/baselines/t2m_baseline/ python calculate_guidance.py --file_type mot --folder_path /home/ubuntu/data/MCS_DATA/LIMO/VQVAE-Generations/mot_visualization/ -
For BIGE
python calculate_guidance.py --file_type mot --folder_path /home/ubuntu/data/MCS_DATA/LIMO/FinalFinalHigh/mot_visualization/
Generate MP4 videos from MOT files.
cd UCSD-OpenCap-Fitness_Dataset/
export DISPLAY=:99.0
python src/opencap_reconstruction_render.py <absolute subject-path> <absolute mot-path> <absolute save-path>We extend our gratitude to The Wu Tsai Human Performance Alliance for their invaluable support and resources. Their dedication to advancing human performance through interdisciplinary research has been instrumental to our project.
This work utilized resources provided by the National Research Platform (NRP) at the University of California, San Diego. The NRP is developed and supported in part by funding from the National Science Foundation under awards 1730158, 1540112, 1541349, 1826967, 2112167, 2100237, and 2120019, along with additional support from community partners.
We also acknowledge the contributions of public code repositories such as text-to-motion, T2M-GPT, MDM, and MotionDiffuse.
Additionally, we appreciate the open-source motion capture systems like OpenCap.