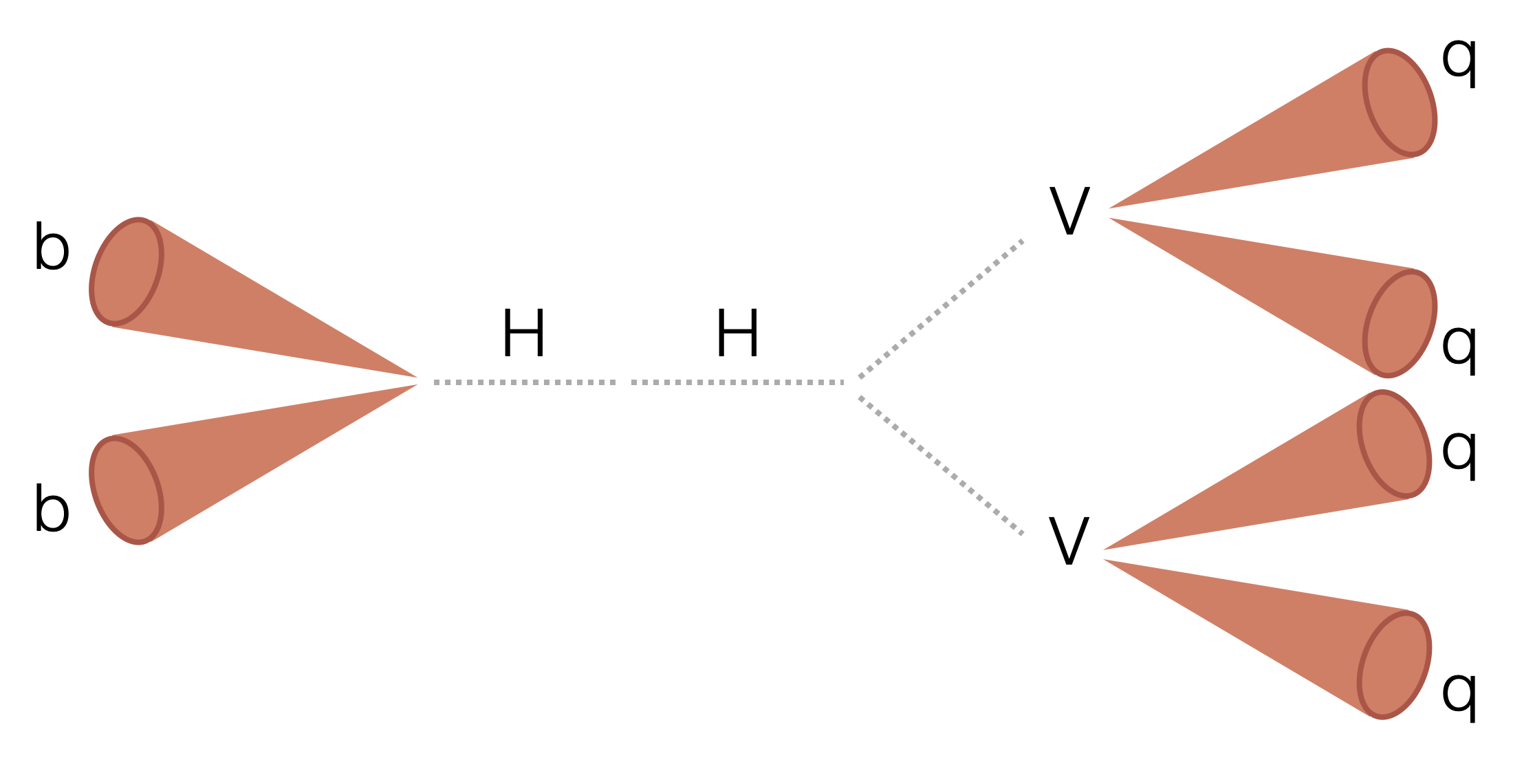
Search for two boosted (high transverse momentum) Higgs bosons (H) decaying to two beauty quarks (b) and two vector bosons (V). The majority of the analysis uses a columnar framework to process input tree-based NanoAOD files using the coffea and scikit-hep Python libraries.
- HHbbVV
First, create a virtual environment (mamba is recommended):
# Download the mamba setup script (change if needed for your machine https://github.com/conda-forge/miniforge#mambaforge)
wget https://github.com/conda-forge/miniforge/releases/latest/download/Mambaforge-Linux-x86_64.sh
# Install: (the mamba directory can end up taking O(1-10GB) so make sure the directory you're using allows that quota)
chmod u+x ./Mambaforge-Linux-x86_64.sh # executable permission
./Mambaforge-Linux-x86_64.sh # follow instructions in the installation
# Clone the repository
git clone https://github.com/rkansal47/HHbbVV.git
cd HHbbVV
# make the environment
mamba env create -f environment.yml
mamba activate bbVV# From inside the HHbbVV repository
# Perform an editable installation
pip install -e .
# for committing to the repository
pre-commit install-
If your default
pythonin your environment is not Python 3, make sure to usepip3andpython3commands instead. -
You may also need to upgrade
pipto perform the editable installation:
python3 -m pip install -e .General note: Coffea-casa is faster and more convenient, however still somewhat experimental so for large of inputs and/or processors which may require heavier cpu/memory usage (e.g. bbVVSkimmer) condor is recommended.
- after following instructions in the link ^ set up an account, open the coffea-casa GUI (https://cmsaf-jh.unl.edu) and create an image
- open
src/runCoffeaCasa.ipynb - import your desired processor, specify it in the
run_uproot_jobfunction, and specify your filelist - run the first three cells
To submit to condor, all you need is python >= 3.7.
For testing locally, it is recommended to use miniconda/mamba (mamba is way faster!):
# Download the setup bash file from here https://github.com/conda-forge/miniforge#mambaforge
# e.g. wget https://github.com/conda-forge/miniforge/releases/latest/download/Mambaforge-Linux-x86_64.sh
# Install: (the mamba directory can end up taking O(1-10GB) so make sure the directory you're using allows that quota)
./Mambaforge-Linux-x86_64.sh # follow instructions in the installation
mamba create -n bbVV python=3.9
mamba activate bbVV
pip install coffea "tritonclient[all]" pyyaml
mamba install -c conda-forge xrootd=5.4.0 # need openssl v1.1 for lxplus and UCSD t2, hence pinning xrootd version.Manually splits up the files into condor jobs.
git clone https://github.com/rkansal47/HHbbVV/
cd HHbbVV
TAG=Aug18_skimmer
# will need python3 (recommended to set up via miniconda)
python src/condor/submit.py --processor skimmer --tag $TAG --files-per-job 20
for i in condor/$TAG/*.jdl; do condor_submit $i; doneAlternatively, can be submitted from a yaml file:
python src/condor/submit_from_yaml.py --year 2017 --processor skimmer --tag $TAG --yaml src/condor/submit_configs/skimmer_inputs_07_24.yamlTo test locally first (recommended), can do e.g.:
mkdir outfiles
python -W ignore src/run.py --starti 0 --endi 1 --year 2017 --processor skimmer --executor iterative --samples HWW --subsamples GluGluToHHTobbVV_node_cHHH1_pn4qApplies a muon pre-selection and accumulates 4D ([Txbb, Th4q, pT, mSD]) yields before and after our triggers.
To test locally:
python -W ignore src/run.py --year 2018 --processor trigger --sample SingleMu2018 --subsamples SingleMuon_Run2018B --starti 0 --endi 1And to submit all:
nohup bash -c 'for i in 2016 2016APV 2017 2018; do python src/condor/submit.py --year $i --tag '"${TAG}"' --processor trigger --submit; done' &> tmp/submitout.txt &Applies pre-selection cuts, runs inference with our new HVV tagger, and saves unbinned branches as parquet files.
Parquet and pickle files will be saved in the eos directory of specified user at
path ~/eos/bbVV/skimmer/<tag>/<sample_name>/<parquet or pickles>. Pickles are
in the format {'nevents': int, 'cutflow': Dict[str, int]}.
To test locally:
python -W ignore src/run.py --processor skimmer --year 2017 --samples HH --subsamples GluGluToHHTobbVV_node_cHHH1 --save-systematics --starti 0 --endi 1or use the src/HHbbVV/bash/run_local.sh to run over files from different
processes.
Or on a specific file(s):
python -W ignore src/run.py --processor skimmer --year 2017 --files $FILE --files-name GluGluToHHTobbVV_node_cHHH1Jobs
nohup python src/condor/submit_from_yaml.py --year 2018 --tag $TAG --processor skimmer --git-branch main --submit --yaml src/condor/submit_configs/skimmer_inputs_24_02_26.yaml &> tmp/submitout.txt &All years:
nohup bash -c 'for year in 2016APV 2016 2017 2018; do python src/condor/submit_from_yaml.py --year $year --tag '"${TAG}"' --processor skimmer --save-systematics --submit --yaml src/condor/submit_configs/skimmer_inputs_23_02_17.yaml; done' &> tmp/submitout.txt &To Submit (if not using the --submit flag)
nohup bash -c 'for i in condor/'"${TAG}"'/*.jdl; do condor_submit $i; done' &> tmp/submitout.txt &Or just signal:
python src/condor/submit.py --year 2017 --tag $TAG --samples HH --subsamples GluGluToHHTobbVV_node_cHHH1 --processor skimmer --submitApplies a loose pre-selection cut, saves ntuples with training inputs.
To test locally:
python -W ignore src/run.py --year 2017 --starti 300 --endi 301 --samples HWWPrivate --subsamples jhu_HHbbWW --processor input --label AK15_H_VV
python -W ignore src/run.py --year 2017 --starti 300 --endi 301 --samples QCD --subsamples QCD_Pt_1000to1400 --processor input --label AK15_QCD --njets 1 --maxchunks 1Jobs:
python src/condor/submit_from_yaml.py --year 2017 --tag $TAG --processor input --save-ak15 --yaml src/condor/submit_configs/training_inputs_07_21.yaml
python src/condor/submit_from_yaml.py --year 2017 --tag $TAG --processor input --yaml src/condor/submit_configs/training_inputs_09_16.yaml --jet AK8To submit add --submit flag.
Applies cuts for a semi-leptonic ttbar control region, as defined for the JMAR W SF and CASE Lund Plane SF measurements to validate Lund plane scale factors.
Lund plane scale factors are calculated for top-matched jets in semi-leptonic ttbar events.
To test locally:
python -W ignore src/run.py --year 2018 --processor ttsfs --sample TTbar --subsamples TTToSemiLeptonic --starti 0 --endi 1Jobs:
nohup python src/condor/submit_from_yaml.py --year 2018 --tag $TAG --processor ttsfs --submit --yaml src/condor/submit_configs/ttsfs_inputs_12_4.yaml &> submitout.txt &Or to submit only the signal:
python src/condor/submit.py --year 2018 --tag $TAG --sample TTbar --subsamples TTToSemiLeptonic --processor ttsfs --submitCheck that all jobs completed by going through output files:
for year in 2016APV 2016 2017 2018; do python src/condor/check_jobs.py --tag $TAG --processor trigger (--submit) --year $year; donenohup version:
(Do condor_q | awk '{ print $9}' | grep -o '[^ ]*\.sh' > running_jobs.txt
first to get a list of jobs which are running.)
nohup bash -c 'for year in 2016APV 2016 2017 2018; do python src/condor/check_jobs.py --year $year --tag '"${TAG}"' --processor skimmer --submit --check-running; done' &> tmp/submitout.txt &Combine all output pickles into one:
for year in 2016APV 2016 2017 2018; do python src/condor/combine_pickles.py --tag $TAG --processor trigger --r --year $year; doneIn `src/HHbbVV/postprocessing':
python BDTPreProcessing.py --data-dir "../../../../data/skimmer/Feb24/" --signal-data-dir "../../../../data/skimmer/Jun10/" --plot-dir "../../../plots/BDTPreProcessing/$TAG/" --year "2017" --bdt-data (--control-plots)python TrainBDT.py --model-dir testBDT --data-path "../../../../data/skimmer/Feb24/bdt_data" --year "all" (--test)Inference-only:
python TrainBDT.py --data-path "../../../../data/skimmer/Feb24/bdt_data" --year "all" --inference-only --model-dir "../../../../data/skimmer/Feb24/23_05_12_multiclass_rem_feats_3"Important: If running on a Mac, make sure to install gnu-getopt first for
bash scripts, see here.
python postprocessing.py --templates --year "2017" --template-dir "templates/$TAG/" --plot-dir "../../../plots/PostProcessing/$TAG/" --data-dir "../../../../data/skimmer/Feb24/" (--resonant --signal-data-dir "" --control-plots)All years (non-resonant):
./bash_scripts/NonresTemplates.sh --tag $TAG # remember to change data_dir as needed!Scan (non-resonant):
./bash_scripts/NonresTemplatesScan.sh --tag $TAG # remember to change data_dir as needed!./bash_scripts/ControlPlot.sh --tag $TAG # w/ resonant and nonresonant samples and all control plot variables in postprocessing.py script by default
./bash_scripts/ControlPlots.sh --tag $TAG --nonresonant --controlplotvars BDTScore --nohem2d # BDT score only
./bash_scripts/MassPlots.sh --tag $TAG # mSD vs mReg plotsRun postprocessing/bash_scripts/BDTPlots.sh from inside
postprocessing folder.
nohup bash_scripts/res_tagger_scan.sh $TAG &> scan.txt &
nohup bash_scripts/res_pt_scan.sh $TAG &> scan.txt &
nohup bash_scripts/res_bias_templates.sh $TAG &> bias.txt &Remember to check output to make sure all years' templates are made!!
Need root==6.22.6 and https://github.com/rkansal47/rhalphalib installed
(pip install -e . --user after cloning the repo).
python3 postprocessing/CreateDatacard.py --templates-dir templates/$TAG --model-name $TAG (--resonant)Or from separate templates for background and signal:
python3 -u postprocessing/CreateDatacard.py --templates-dir "/eos/uscms/store/user/rkansal/bbVV/templates/23Apr30Scan/txbb_HP_thww_0.96" \
--sig-separate --resonant --model-name $sample --sig-sample $sampleScan (non-resonant):
for txbb_wp in "LP" "MP" "HP"; do for bdt_wp in 0.994 0.99 0.96 0.9 0.8 0.6 0.4; do python3 -u postprocessing/CreateDatacard.py --templates-dir templates/23May13NonresScan/txbb_${txbb_wp}_bdt_${bdt_wp} --model-name 23May13NonresScan/txbb_${txbb_wp}_bdt_${bdt_wp} --no-do-jshifts --nTF 0; done; doneDatacards with different orders of TFs for F-tests:
Use the src/HHbbVV/combine/F_test_res.sh script.
Datacards (and background-only fits) for bias tests:
src/HHbbVV/combine/biastests.sh --cardstag $cardstag --templatestag $templatestag
python PlotFits.py --fit-file "cards/test_tied_stats/fitDiagnosticsBlindedBkgOnly.root" --plots-dir "../../../plots/PostFit/09_02/"Warning: this should be done outside of your conda/mamba environment!
source /cvmfs/cms.cern.ch/cmsset_default.sh
cmsrel CMSSW_11_3_4
cd CMSSW_11_3_4/src
cmsenv
# git clone -b main https://github.com/rkansal47/HiggsAnalysis-CombinedLimit.git HiggsAnalysis/CombinedLimit
# float regex PR was merged so we should be able to switch to the main branch now:
git clone -b v9.2.0 https://github.com/cms-analysis/HiggsAnalysis-CombinedLimit.git HiggsAnalysis/CombinedLimit
git clone -b v2.0.0 https://github.com/cms-analysis/CombineHarvester.git CombineHarvester
# Important: this scram has to be run from src dir
scramv1 b clean; scramv1 bTo create datacards, you need to use the same cmsenv as above + these packages:
pip3 install --upgrade pip
pip3 install rhalphalib
cd /path/to/your/local/HHbbVV/repo
pip3 install -e .I also add this to my .bashrc for convenience:
export PATH="$PATH:/uscms_data/d1/rkansal/HHbbVV/src/HHbbVV/combine"
csubmit() {
local file=$1; shift;
python3 "/uscms_data/d1/rkansal/HHbbVV/src/HHbbVV/combine/submit/submit_${file}.py" "$@"
}
All via the below script, with a bunch of options (see script):
run_blinded.sh --workspace --bfit --limitsThis will take 5-10 minutes for 100 toys will take forever for more than >>100!.
# automatically make workspaces and do the background-only fit for orders 0 - 3
run_ftest_nonres.sh --sample HHbbVV --cardstag 24Apr10ggFMP9965 --templatestag 24Apr9ggFMP9965 # -dl for saving shapes and limits
# run f-test for desired order
run_ftest_nonres.sh --sample HHbbVV --cardstag 24Apr10ggFMP9965 --goftoys --ffits --numtoys 100 --seed 444 --order 0VBF:
# automatically make workspaces and do the background-only fit for orders 0 - 3
run_ftest_nonres.sh --sample qqHH_CV_1_C2V_0_kl_1_HHbbVV --templatestag 24Apr8VBFHP999 --cardstag 24Apr8VBFHP999 -dl # -dl for saving shapes and limits
# run f-test for desired order
run_ftest_nonres.sh --sample qqHH_CV_1_C2V_0_kl_1_HHbbVV --templatestag 24Apr8VBFHP999 --cardstag 24Apr8VBFHP999 ---goftoys --ffits --numtoys 100 --seed 444 --order 1Condor is needed for >100 toys or resonant, see below.
Can run over all the resonant signals (default) or scan working points for a
subset of signals (--scan)
csubmit cards --test --scan --resonant --templates-dir 23Apr30ScanGenerate toys and fits for F-tests (after making cards and b-only fits for the testing order AND testing order + 1!)
Nonresonant:
csubmit ftest --tag 24Apr9 --cards-tag 24Apr9VBFHP999 --low1 0 --no-resonant --num-jobs 5 # 500 toysResonant:
csubmit ftest --tag 23May2 --cards-tag 23May2 --low1 0 --low2 0 # 1000 toyscsubmit impacts --tag 23May2 (--local [if you want to run them locally])This was also output a script to collect all the impacts after the jobs finish.
For resonant, use scripts inside the src/HHbbVV/combine/ directory and run
from one level above the sample datacard folders (e.g.
/uscms/home/rkansal/hhcombine/cards/biastests/23Jul17ResClipTFScale1).
Setting up the datacards, assuming templates have already been made (see templates section), e.g.:
setup_bias.sh -r --scale 1 --cardstag 23Sep14_hww0.6_pt_400_350 --templatestag 23Sep14_thww0.6_pt_400_350Submitting 1000 toys for each sample and r value + more toys for samples with
high fit failure rates:
submit_bias_res_loop.sh $seed $TAGMoving the outputs to a bias/$TAG dir after (uses the last digit of the seed -
so make sure different runs use different last digits!):
mv_bias_outputs.sh [last-digit-of-seed] $TAGTo run bash scripts with the getopt command (e.g. run_blinded.sh,
ControlPlots.sh) on Macs:
# install gnu-getopt with Homebrew
brew install gnu-getopt
# add to path
sudo echo 'export PATH="/usr/local/opt/gnu-getopt/bin:$PATH"' >> ~/.zsh_profile
source ~/.zsh_profile(krsync) (you may need to install rsync in
the PRP pod first if it's been restarted)
cd ~/eos/bbVV/input/${TAG}_${JETS}/2017
mkdir ../copy_logs
for i in *; do echo $i && sleep 3 && (nohup sh -c "krsync -av --progress --stats $i/root/ $HWWTAGGERDEP_POD:/hwwtaggervol/training/$FOLDER/$i" &> ../copy_logs/$i.txt &) donefor sample in 'NMSSM_XToYHTo2W2BTo4Q2B_MX-900_MY-80' 'NMSSM_XToYHTo2W2BTo4Q2B_MX-1200_MY-190' 'NMSSM_XToYHTo2W2BTo4Q2B_MX-2000_MY-125' 'NMSSM_XToYHTo2W2BTo4Q2B_MX-3000_MY-250' 'NMSSM_XToYHTo2W2BTo4Q2B_MX-4000_MY-150'; do for year in 2016APV 2016 2017 2018; do rsync -avP [email protected]:eos/bbVV/skimmer/Apr11/$year/$sample data/skimmer/Apr11/$year/; done; donecondor_q | awk '{ print $9}' | grep -o '[^ ]*\.sh'Kill each task:
dataset=SingleMuon
for i in {B..F}; do crab kill -d crab/pfnano_v2_3/crab_pfnano_v2_3_2016_${dataset}_Run2016${i}*HIPM; done
for i in {F..H}; do crab kill -d crab/pfnano_v2_3/crab_pfnano_v2_3_2016_${dataset}_Run2016$i; done
for i in {B..F}; do crab kill -d crab/pfnano_v2_3/crab_pfnano_v2_3_2017_${dataset}_Run2017$i; done
for i in {A..D}; do crab kill -d crab/pfnano_v2_3/crab_pfnano_v2_3_2018_${dataset}_Run2018$i; doneGet a crab report for each task:
for i in {B..F}; do crab report -d crab/pfnano_v2_3/crab_pfnano_v2_3_2016_${dataset}_Run2016${i}*HIPM; done
for i in {F..H}; do crab report -d crab/pfnano_v2_3/crab_pfnano_v2_3_2016_${dataset}_Run2016$i; done
for i in {B..F}; do crab report -d crab/pfnano_v2_3/crab_pfnano_v2_3_2017_${dataset}_Run2017$i; done
for i in {A..D}; do crab report -d crab/pfnano_v2_3/crab_pfnano_v2_3_2018_${dataset}_Run2018$i; doneCombine the lumis to process:
mkdir -p recovery/$dataset/
# shopt extglob
# jq -s 'reduce .[] as $item ({}; . * $item)' crab/pfnano_v2_3/crab_pfnano_v2_3_2016_${dataset}_*HIPM/results/notFinishedLumis.json > recovery/$dataset/2016APVNotFinishedLumis.json
for year in 2016 2017 2018; do jq -s 'reduce .[] as $item ({}; . * $item)' crab/pfnano_v2_3/crab_pfnano_v2_3_${year}_${dataset}_*/results/notFinishedLumis.json > recovery/$dataset/${year}NotFinishedLumis.json; doneFinally, add these as a lumimask for the recovery task.