This is a fork of the original CoMut package from the Van Allen Lab to update source code. To use, git clone this repository:
git clone [email protected]:getzlab/van_allen_comut.git
Enter the cloned repository via
cd van_allen_comut
and install with
pip install -e .
preferably within a conda environment.
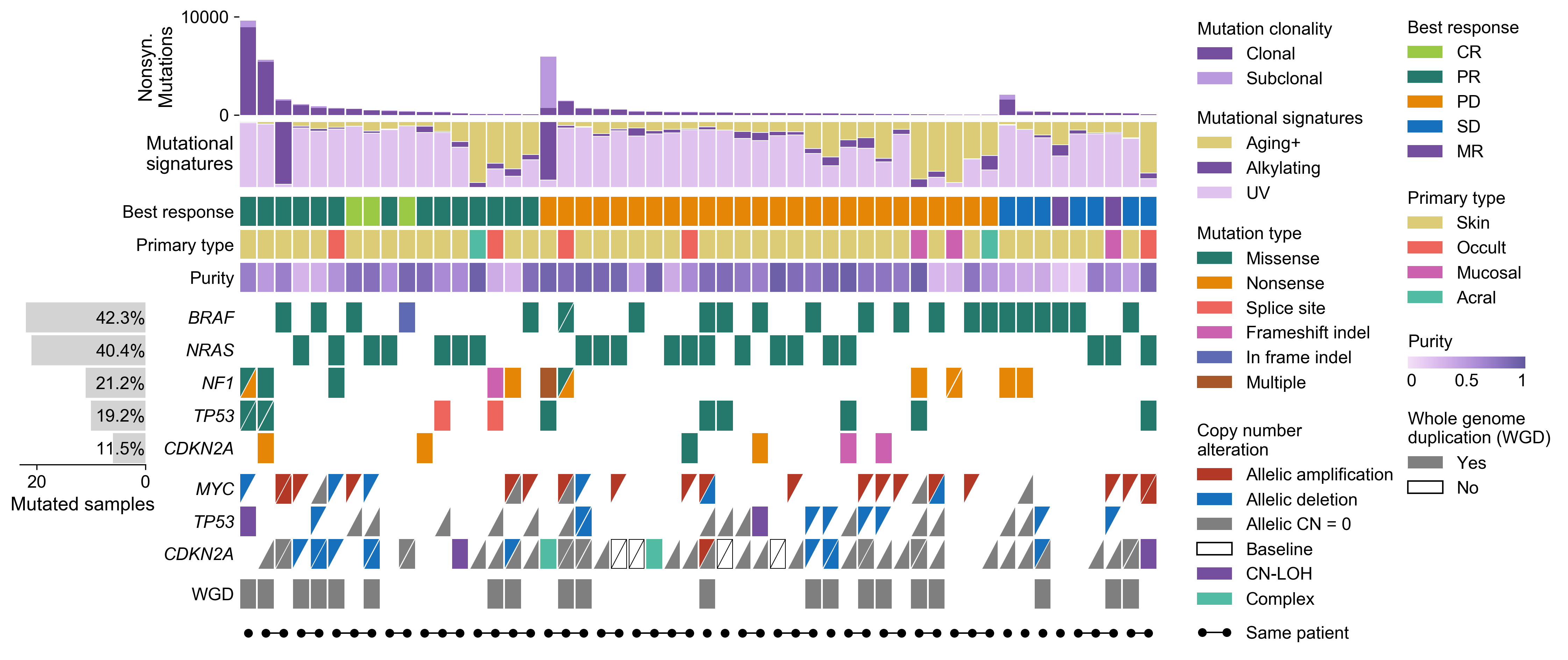
CoMut is a Python library for creating comutation plots to visualize genomic and phenotypic information.
CoMut is available on pypi here and can be installed via pip
pip install comut
For those who do not want to install Python or other packages, there is a Google Colab notebook where you can simply upload a MAF file and run the notebook to make a basic comutation plot. This file is also available as a jupyter notebook for local use.
CoMut is now published here - https://academic.oup.com/bioinformatics/article/doi/10.1093/bioinformatics/btaa554/5851837. If you use CoMut in a paper, please cite:
Crowdis, J., He, M.X., Reardon, B. & Van Allen, E. M. CoMut: Visualizing integrated molecular information with comutation plots. Bioinformatics (2020). doi:10.1093/bioinformatics/btaa554
There is also a Documentation notebook that provides documentation for CoMut. It describes the fundamentals of creating comutation plots and provides the code used to generate the comut above.
If you would like to report a bug or request a feature, please do so using the issues page
CoMut runs on python 3.6 or later. CoMut requires the following packages as dependencies (they will be installed along with CoMut if using pip)
numpy>=1.18.1
pandas>=0.25.3
palettable>=3.3.0
matplotlib>=3.3.1
0.0.3 - No code is changed, description updated for public release
0.0.2 - Introduce compatability for Python 3.6
0.0.1 - Initial release