-
Notifications
You must be signed in to change notification settings - Fork 8
/
generate_README.sh
executable file
·248 lines (216 loc) · 8.85 KB
/
generate_README.sh
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
204
205
206
207
208
209
210
211
212
213
214
215
216
217
218
219
220
221
222
223
224
225
226
227
228
229
230
231
232
233
234
235
236
237
238
239
240
241
242
243
244
245
246
247
248
#!/usr/bin/env zsh
# -*- coding: UTF8 -*-
#############################################################################
# Authors: Vincent Mallet, Guillaume Bouvier #
# Copyright (c) 2021 Institut Pasteur #
# #
# #
# Redistribution and use in source and binary forms, with or without #
# modification, are permitted provided that the following conditions #
# are met: #
# #
# 1. Redistributions of source code must retain the above copyright #
# notice, this list of conditions and the following disclaimer. #
# 2. Redistributions in binary form must reproduce the above copyright #
# notice, this list of conditions and the following disclaimer in the #
# documentation and/or other materials provided with the distribution. #
# 3. Neither the name of the copyright holder nor the names of its #
# contributors may be used to endorse or promote products derived from #
# this software without specific prior written permission. #
# #
# THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS #
# "AS IS" AND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT #
# LIMITED TO, THE IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR #
# A PARTICULAR PURPOSE ARE DISCLAIMED. IN NO EVENT SHALL THE COPYRIGHT #
# HOLDER OR CONTRIBUTORS BE LIABLE FOR ANY DIRECT, INDIRECT, INCIDENTAL, #
# SPECIAL, EXEMPLARY, OR CONSEQUENTIAL DAMAGES (INCLUDING, BUT NOT #
# LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR SERVICES; LOSS OF USE, #
# DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER CAUSED AND ON ANY #
# THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY, OR TORT #
# (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE #
# OF THIS SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE. #
# #
# This program is free software: you can redistribute it and/or modify #
# #
#############################################################################
func runcmd() {
OUTPUT=$(eval $1)
echo "\`\`\`"
echo "$ $1\n"
echo "$OUTPUT"
echo "\`\`\`"
}
func runcmd_cut() {
OUTPUT=$(eval $1)
echo "\`\`\`"
echo "$ $1\n"
echo "$OUTPUT" | head -3
echo "[...]"
echo "$OUTPUT" | tail -3
echo "\`\`\`"
}
func runcmd_null() {
OUTPUT=$(eval $1)
echo "\`\`\`bash"
echo "$1\n"
echo "\`\`\`"
}
[ -d figures ] && rm -r figures
cat << EOF
# Self-Organizing Map
PyTorch implementation of a Self-Organizing Map.
The implementation makes possible the use of a GPU if available for faster computations.
It follows the scikit package semantics for training and usage of the model.
It also includes runnable scripts to avoid coding.
Example of a MD clustering using \`quicksom\`:
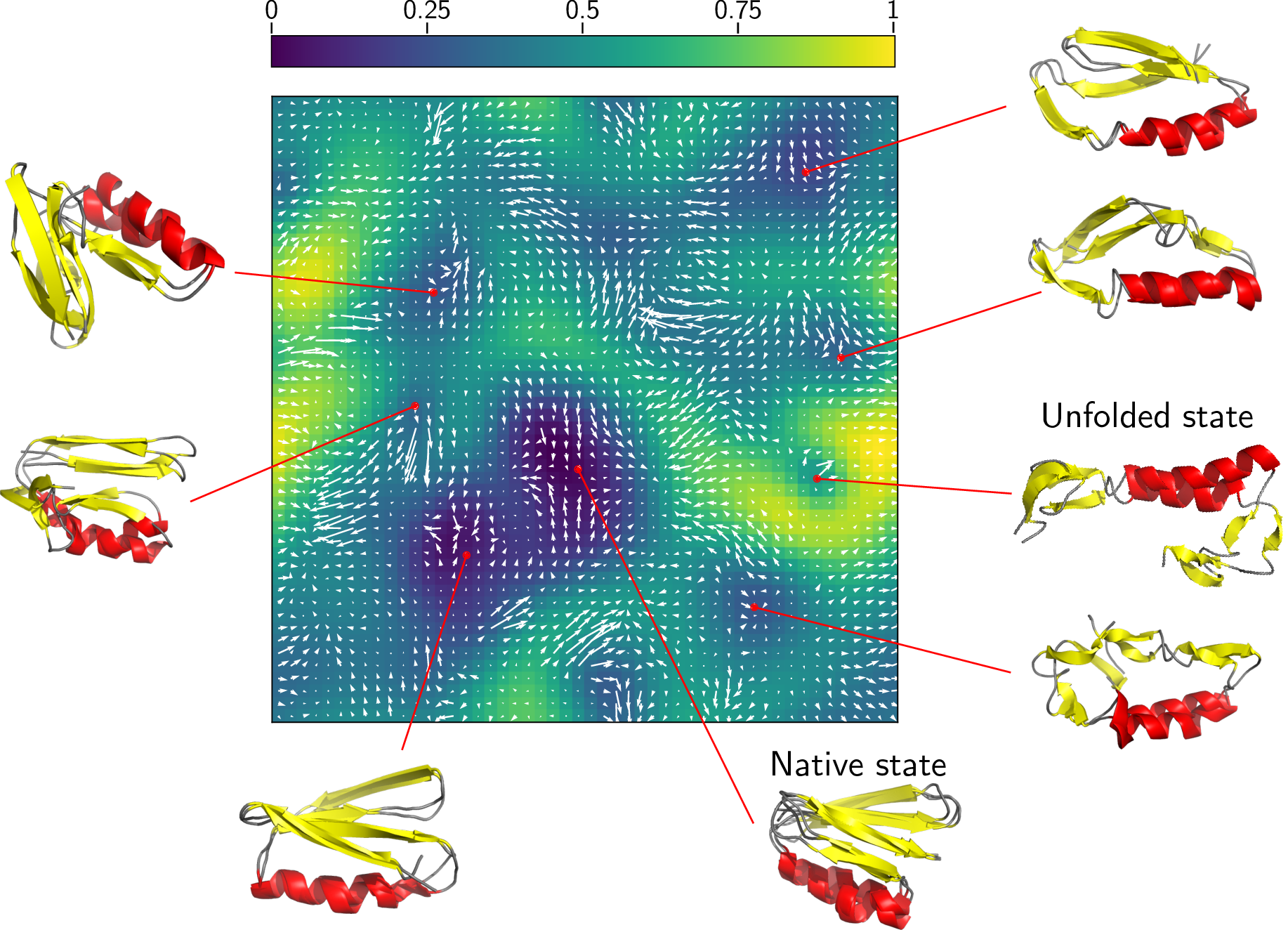
EOF
cat << EOF
### Requirements and setup
The SOM object requires PyTorch installed.
It has dependencies in numpy, scipy and scikit-learn and scikit-image.
The MD application requires pymol to load the trajectory that is not included in the dependencies
To set up the project, we suggest using conda environments.
Install [PyTorch](https://pytorch.org/get-started/locally/) and run :
\`\`\`
pip install quicksom
\`\`\`
EOF
cat << EOF
### SOM object interface
The SOM object can be created using any grid size, with a optional periodic topology.
One can also choose optimization parameters such as the number of epochs to train or the batch size
To use it, we include three scripts to :
- fit a SOM
- to optionally build the clusters manually with a gui
- to predict cluster affectations for new data points
EOF
runcmd "quicksom_fit -h"
runcmd "quicksom_gui -h"
runcmd "quicksom_predict -h"
cat << EOF
The SOM object is also importable from python scripts to use
directly in your analysis pipelines :
EOF
cat << EOF
\`\`\`python
import pickle
import numpy
import torch
from som import SOM
# Get data
device = 'cuda' if torch.cuda.is_available() else 'cpu'
X = numpy.load('contact_desc.npy')
X = torch.from_numpy(X)
X = X.float()
X = X.to(device)
# Create SOM object and train it, then dump it as a pickle object
m, n = 100, 100
dim = X.shape[1]
niter = 5
batch_size = 100
som = SOM(m, n, dim, niter=niter, device=device)
learning_error = som.fit(X, batch_size=batch_size)
som.to_device('cpu')
pickle.dump(som, open('som.pickle', 'wb'))
# Usage on the input data, predicted_clusts is an array of length n_samples with clusters affectations
som = pickle.load(open('som.pickle', 'rb'))
som.to_device(device)
predicted_clusts, errors = som.predict_cluster(X)
\`\`\`
EOF
cat << EOF
### SOM training on molecular dynamics (MD) trajectories
#### Scripts and extra dependencies:
- \`dcd2npy\`: [Pymol](https://anaconda.org/schrodinger/pymol)
- \`mdx\`: [Pymol](https://anaconda.org/schrodinger/pymol), [pymol-psico](https://github.com/speleo3/pymol-psico)
To set these dependencies up using conda, just type :
\`\`\`
conda install -c schrodinger pymol pymol-psico
\`\`\`
The SOM algorithm can efficiently map MD trajectories for analysis and clustering purposes.
The script \`dcd2npy\` can be used to select a subset of atoms from a trajectory in \`dcd\` format,
align it and save the selection as a \`npy\` file that can be handled by the command \`quicksom_fit\`.
EOF
runcmd "dcd2npy -h"
cat << EOF
The following commands can be applied for a MD clustering.
#### Create a npy file with atomic coordinates of C-alpha:
EOF
runcmd_cut "dcd2npy --pdb data/2lj5.pdb --dcd data/2lj5.dcd --select 'name CA'"
cat << EOF
#### Fit the SOM:
EOF
if [ -f data/som_2lj5.p ]; then
cat << EOF
\`\`\`
$ quicksom_fit -i data/2lj5.npy -o data/som_2lj5.p --n_iter 100 --batch_size 50 --periodic --alpha 0.5
1/100: 50/301 | alpha: 0.500000 | sigma: 25.000000 | error: 397.090729 | time 0.387760
4/100: 150/301 | alpha: 0.483333 | sigma: 24.166667 | error: 8.836357 | time 5.738029
7/100: 250/301 | alpha: 0.466667 | sigma: 23.333333 | error: 8.722509 | time 11.213565
[...]
91/100: 50/301 | alpha: 0.050000 | sigma: 2.500000 | error: 5.658005 | time 137.348755
94/100: 150/301 | alpha: 0.033333 | sigma: 1.666667 | error: 5.373021 | time 142.033695
97/100: 250/301 | alpha: 0.016667 | sigma: 0.833333 | error: 5.855451 | time 147.203326
\`\`\`
EOF
else
runcmd_cut "quicksom_fit -i data/2lj5.npy -o data/som_2lj5.p --n_iter 100 --batch_size 50 --periodic --alpha 0.5"
fi
cat << EOF
#### Analysis and clustering of the map using \`quicksom_gui\`:
\`\`\`bash
quicksom_gui -i data/som_2lj5.p
\`\`\`
### Analysis of MD trajectories with this SOM
We now have a trained SOM and we can use several functionalities such as clustering input data points and grouping them
into separate dcd files, creating a dcd with one centroid per fram or plotting of the U-Matrix and its flow.
#### Cluster assignment of input data points:
EOF
runcmd_null "quicksom_predict -i data/2lj5.npy -o data/2lj5 -s data/som_2lj5.p"
cat << EOF
This command generates 3 files:
EOF
runcmd "ls data/2lj5_*.txt"
cat << EOF
containing the data:
- Best Matching Unit with error for each data point
- Cluster assignment
- Assignment for each SOM cell of the closest data point (BMU with minimal error). \`-1\` means no assignment
EOF
runcmd "head -3 data/2lj5_bmus.txt"
runcmd "head -3 data/2lj5_clusters.txt"
cat << EOF
#### Cluster extractions from the input \`dcd\` using the \`quicksom_extract\` tool:
EOF
runcmd 'quicksom_extract -h'
runcmd_null 'quicksom_extract -p data/2lj5.pdb -t data/2lj5.dcd -c data/2lj5_clusters.txt'
runcmd "ls -v data/cluster_*.dcd"
cat << EOF
#### Extraction of the SOM centroids from the input \`dcd\`
EOF
runcmd_null 'grep -v "\-1" data/2lj5_codebook.txt > _codebook.txt
mdx --top data/2lj5.pdb --traj data/2lj5.dcd --fframes _codebook.txt --out data/centroids.dcd
rm _codebook.txt'
cat << EOF
#### Plotting the U-matrix:
EOF
runcmd_null "python3 -c 'import pickle
import matplotlib.pyplot as plt
som=pickle.load(open(\"data/som_2lj5.p\", \"rb\"))
plt.matshow(som.umat)
plt.savefig(\"data/umat_2lj5.png\")
'"
cat << EOF
#### Flow analysis
The flow of the trajectory can be projected onto the U-matrix using the following command:
EOF
runcmd "quicksom_flow -h"
cat << EOF
With this toy example, we get the following plot:
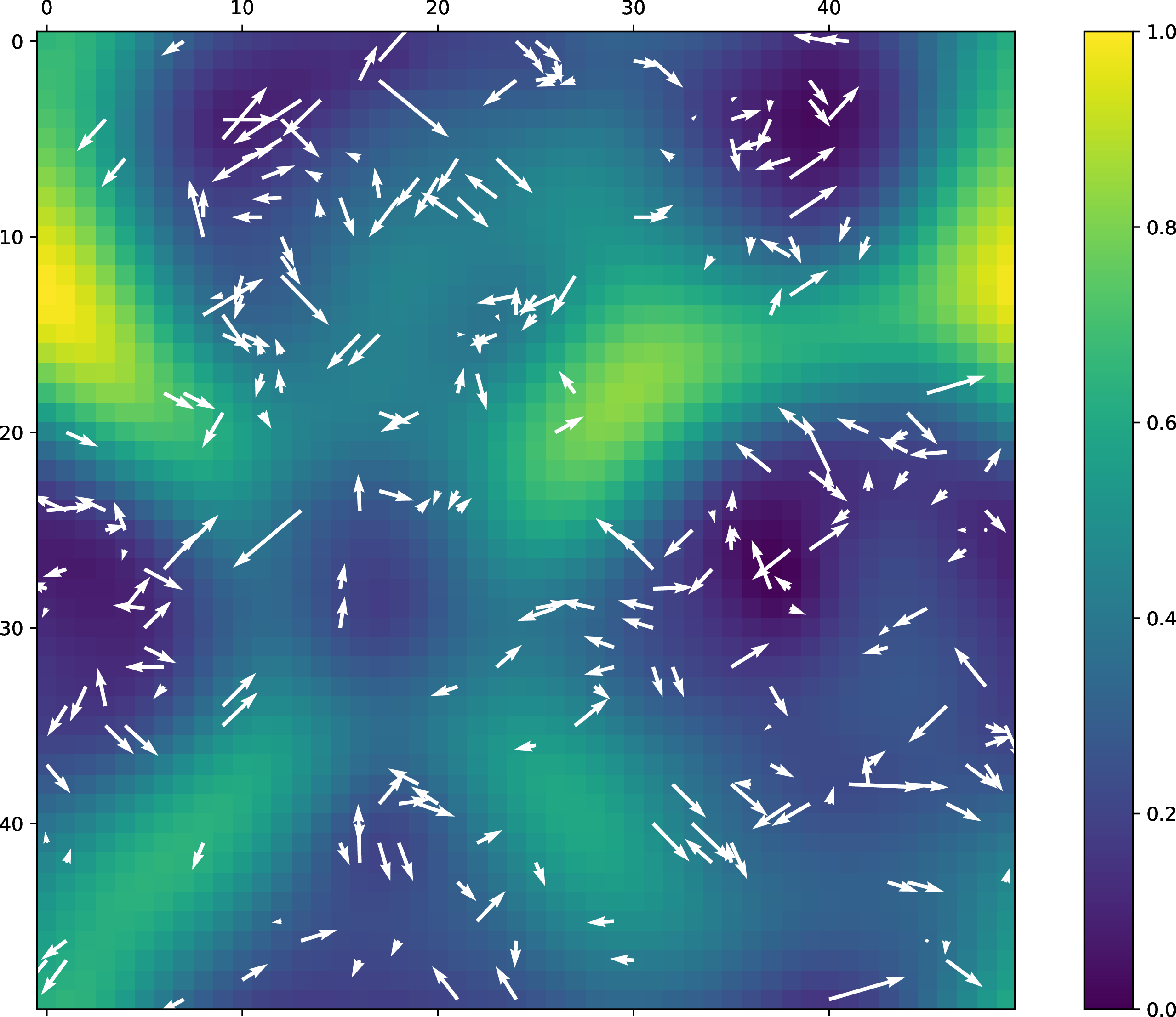
EOF
cat << EOF
#### Data projection
EOF
runcmd "quicksom_project -h"