diff --git a/.DS_Store b/.DS_Store
deleted file mode 100644
index 03ce2149..00000000
Binary files a/.DS_Store and /dev/null differ
diff --git a/.github/FUNDING.yml b/.github/FUNDING.yml
new file mode 100644
index 00000000..d68b17e1
--- /dev/null
+++ b/.github/FUNDING.yml
@@ -0,0 +1,12 @@
+# These are supported funding model platforms
+
+github: # Replace with up to 4 GitHub Sponsors-enabled usernames e.g., [user1, user2]
+patreon: # Replace with a single Patreon username
+open_collective: # Replace with a single Open Collective username
+ko_fi: # Replace with a single Ko-fi username
+tidelift: # Replace with a single Tidelift platform-name/package-name e.g., npm/babel
+community_bridge: # Replace with a single Community Bridge project-name e.g., cloud-foundry
+liberapay: # Replace with a single Liberapay username
+issuehunt: # Replace with a single IssueHunt username
+otechie: # Replace with a single Otechie username
+custom: ['https://www.paypal.me/Udayraj123/','https://www.buymeacoffee.com/Udayraj123']
diff --git a/.github/ISSUE_TEMPLATE/bug_report.md b/.github/ISSUE_TEMPLATE/bug_report.md
new file mode 100644
index 00000000..408ab293
--- /dev/null
+++ b/.github/ISSUE_TEMPLATE/bug_report.md
@@ -0,0 +1,34 @@
+---
+name: Bug report
+about: Create a report to help us improve
+title: "[Bug]"
+labels: ''
+assignees: ''
+
+---
+
+**Describe the bug**
+A clear and concise description of what the bug is.
+
+**To Reproduce**
+Steps to reproduce the behavior:
+1. Use sample '...'
+2. Command(s) used '....'
+3. See error
+
+**Expected behavior**
+A clear and concise description of what you expected to happen.
+
+**Screenshots**
+If applicable, add screenshots to help explain your problem.
+
+**Desktop (please complete the following information):**
+ - OS: [e.g. MacOS, Linux, Windows]
+ - Python version
+ - OpenCV version
+
+
+**Additional context**
+Add any other context about the problem here.
+
+Error Stack trace. Sample images used, etc
diff --git a/.github/ISSUE_TEMPLATE/feature_request.md b/.github/ISSUE_TEMPLATE/feature_request.md
new file mode 100644
index 00000000..604341ba
--- /dev/null
+++ b/.github/ISSUE_TEMPLATE/feature_request.md
@@ -0,0 +1,20 @@
+---
+name: Feature request
+about: Suggest an idea/enhancement for this project
+title: "[Feature]"
+labels: enhancement
+assignees: ''
+
+---
+
+**Is your feature request related to a problem? Please describe.**
+A clear and concise description of what the problem is. Ex. I think implementing [...] will help everyone.
+
+**Describe the solution you'd like**
+A clear and concise description of what you want to happen.
+
+**Describe alternatives you've considered**
+A clear and concise description of any alternative solutions or features you've considered.
+
+**Additional context**
+Add any other context or screenshots about the feature request here.
diff --git a/.github/pre-commit.yml b/.github/pre-commit.yml
new file mode 100644
index 00000000..d6b9d9a6
--- /dev/null
+++ b/.github/pre-commit.yml
@@ -0,0 +1,13 @@
+name: Pre-Commit Hook
+
+on: [push, pull_request]
+
+jobs:
+ pre-commit:
+ runs-on: ubuntu-latest
+ steps:
+ - uses: actions/checkout@v3
+ - uses: actions/setup-python@v3
+ with:
+ python-version: '3.11'
+ - uses: pre-commit/action@v3.0.0
diff --git a/.gitignore b/.gitignore
index 2778cf2c..a7d4046b 100644
--- a/.gitignore
+++ b/.gitignore
@@ -1,12 +1,14 @@
-**/__pycache__
-**/CheckedOMRs
-**/ignore
-**/.DS_Store
-OMRChecker.wiki/
+# Any directory starting with a dot
+**/\.*/
+# Except .github
+!.github/
+
# Everything in inputs/ and outputs/
inputs/*
outputs/*
-# Except *.json and OMR_Files/
-# !inputs/OMR_Files/
-# !inputs/*.json
-# !inputs/omr_marker.jpg
\ No newline at end of file
+
+# Misc
+**/.DS_Store
+**/__pycache__
+venv/
+OMRChecker.wiki/
diff --git a/.pre-commit-config.yaml b/.pre-commit-config.yaml
new file mode 100644
index 00000000..2daa2af8
--- /dev/null
+++ b/.pre-commit-config.yaml
@@ -0,0 +1,59 @@
+exclude: "__snapshots__/.*$"
+default_install_hook_types: [pre-commit, pre-push]
+repos:
+ - repo: https://github.com/pre-commit/pre-commit-hooks
+ rev: v4.4.0
+ hooks:
+ - id: check-yaml
+ stages: [commit]
+ - id: check-added-large-files
+ args: ['--maxkb=300']
+ fail_fast: false
+ stages: [commit]
+ - id: pretty-format-json
+ args: ['--autofix', '--no-sort-keys']
+ - id: end-of-file-fixer
+ exclude_types: ["csv", "json"]
+ stages: [commit]
+ - id: trailing-whitespace
+ stages: [commit]
+ - repo: https://github.com/pycqa/isort
+ rev: 5.12.0
+ hooks:
+ - id: isort
+ args: ["--profile", "black"]
+ stages: [commit]
+ - repo: https://github.com/psf/black
+ rev: 23.3.0
+ hooks:
+ - id: black
+ fail_fast: true
+ stages: [commit]
+ - repo: https://github.com/pycqa/flake8
+ rev: 6.0.0
+ hooks:
+ - id: flake8
+ args:
+ - "--ignore=E501,W503,E203,E741,F541" # Line too long, Line break occurred before a binary operator, Whitespace before ':'
+ fail_fast: true
+ stages: [commit]
+ - repo: local
+ hooks:
+ - id: pytest-on-commit
+ name: Running single sample test
+ entry: python3 -m pytest -rfpsxEX --disable-warnings --verbose -k sample1
+ language: system
+ pass_filenames: false
+ always_run: true
+ fail_fast: true
+ stages: [commit]
+ - repo: local
+ hooks:
+ - id: pytest-on-push
+ name: Running all tests before push...
+ entry: python3 -m pytest -rfpsxEX --disable-warnings --verbose --durations=3
+ language: system
+ pass_filenames: false
+ always_run: true
+ fail_fast: true
+ stages: [push]
diff --git a/.pylintrc b/.pylintrc
new file mode 100644
index 00000000..405ce2e7
--- /dev/null
+++ b/.pylintrc
@@ -0,0 +1,43 @@
+[BASIC]
+# Regular expression matching correct variable names. Overrides variable-naming-style.
+# snake_case with single letter regex -
+variable-rgx=[a-z0-9_]{1,30}$
+
+# Good variable names which should always be accepted, separated by a comma.
+good-names=x,y,pt
+
+[MESSAGES CONTROL]
+
+# Disable the message, report, category or checker with the given id(s). You
+# can either give multiple identifiers separated by comma (,) or put this
+# option multiple times (only on the command line, not in the configuration
+# file where it should appear only once). You can also use "--disable=all" to
+# disable everything first and then reenable specific checks. For example, if
+# you want to run only the similarities checker, you can use "--disable=all
+# --enable=similarities". If you want to run only the classes checker, but have
+# no Warning level messages displayed, use "--disable=all --enable=classes
+# --disable=W".
+disable=import-error,
+ unresolved-import,
+ too-few-public-methods,
+ missing-docstring,
+ relative-beyond-top-level,
+ too-many-instance-attributes,
+ bad-continuation,
+ no-member
+
+# Note: bad-continuation is a false positive showing bug in pylint
+# https://github.com/psf/black/issues/48
+
+
+[REPORTS]
+# Set the output format. Available formats are text, parseable, colorized, json
+# and msvs (visual studio). You can also give a reporter class, e.g.
+# mypackage.mymodule.MyReporterClass.
+output-format=text
+
+# Tells whether to display a full report or only the messages.
+reports=no
+
+# Activate the evaluation score.
+score=yes
diff --git a/CODE_OF_CONDUCT.md b/CODE_OF_CONDUCT.md
new file mode 100644
index 00000000..5a01f7a6
--- /dev/null
+++ b/CODE_OF_CONDUCT.md
@@ -0,0 +1,133 @@
+
+# Contributor Covenant Code of Conduct
+
+## Our Pledge
+
+We as members, contributors, and leaders pledge to make participation in our
+community a harassment-free experience for everyone, regardless of age, body
+size, visible or invisible disability, ethnicity, sex characteristics, gender
+identity and expression, level of experience, education, socio-economic status,
+nationality, personal appearance, race, caste, color, religion, or sexual
+identity and orientation.
+
+We pledge to act and interact in ways that contribute to an open, welcoming,
+diverse, inclusive, and healthy community.
+
+## Our Standards
+
+Examples of behavior that contributes to a positive environment for our
+community include:
+
+* Demonstrating empathy and kindness toward other people
+* Being respectful of differing opinions, viewpoints, and experiences
+* Giving and gracefully accepting constructive feedback
+* Accepting responsibility and apologizing to those affected by our mistakes,
+ and learning from the experience
+* Focusing on what is best not just for us as individuals, but for the overall
+ community
+
+Examples of unacceptable behavior include:
+
+* The use of sexualized language or imagery, and sexual attention or advances of
+ any kind
+* Trolling, insulting or derogatory comments, and personal or political attacks
+* Public or private harassment
+* Publishing others' private information, such as a physical or email address,
+ without their explicit permission
+* Other conduct which could reasonably be considered inappropriate in a
+ professional setting
+
+## Enforcement Responsibilities
+
+Community leaders are responsible for clarifying and enforcing our standards of
+acceptable behavior and will take appropriate and fair corrective action in
+response to any behavior that they deem inappropriate, threatening, offensive,
+or harmful.
+
+Community leaders have the right and responsibility to remove, edit, or reject
+comments, commits, code, wiki edits, issues, and other contributions that are
+not aligned to this Code of Conduct, and will communicate reasons for moderation
+decisions when appropriate.
+
+## Scope
+
+This Code of Conduct applies within all community spaces, and also applies when
+an individual is officially representing the community in public spaces.
+Examples of representing our community include using an official email address,
+posting via an official social media account, or acting as an appointed
+representative at an online or offline event.
+
+## Enforcement
+
+Instances of abusive, harassing, or otherwise unacceptable behavior may be
+reported to the community leaders responsible for enforcement at
+[INSERT CONTACT METHOD].
+All complaints will be reviewed and investigated promptly and fairly.
+
+All community leaders are obligated to respect the privacy and security of the
+reporter of any incident.
+
+## Enforcement Guidelines
+
+Community leaders will follow these Community Impact Guidelines in determining
+the consequences for any action they deem in violation of this Code of Conduct:
+
+### 1. Correction
+
+**Community Impact**: Use of inappropriate language or other behavior deemed
+unprofessional or unwelcome in the community.
+
+**Consequence**: A private, written warning from community leaders, providing
+clarity around the nature of the violation and an explanation of why the
+behavior was inappropriate. A public apology may be requested.
+
+### 2. Warning
+
+**Community Impact**: A violation through a single incident or series of
+actions.
+
+**Consequence**: A warning with consequences for continued behavior. No
+interaction with the people involved, including unsolicited interaction with
+those enforcing the Code of Conduct, for a specified period of time. This
+includes avoiding interactions in community spaces as well as external channels
+like social media. Violating these terms may lead to a temporary or permanent
+ban.
+
+### 3. Temporary Ban
+
+**Community Impact**: A serious violation of community standards, including
+sustained inappropriate behavior.
+
+**Consequence**: A temporary ban from any sort of interaction or public
+communication with the community for a specified period of time. No public or
+private interaction with the people involved, including unsolicited interaction
+with those enforcing the Code of Conduct, is allowed during this period.
+Violating these terms may lead to a permanent ban.
+
+### 4. Permanent Ban
+
+**Community Impact**: Demonstrating a pattern of violation of community
+standards, including sustained inappropriate behavior, harassment of an
+individual, or aggression toward or disparagement of classes of individuals.
+
+**Consequence**: A permanent ban from any sort of public interaction within the
+community.
+
+## Attribution
+
+This Code of Conduct is adapted from the [Contributor Covenant][homepage],
+version 2.1, available at
+[https://www.contributor-covenant.org/version/2/1/code_of_conduct.html][v2.1].
+
+Community Impact Guidelines were inspired by
+[Mozilla's code of conduct enforcement ladder][Mozilla CoC].
+
+For answers to common questions about this code of conduct, see the FAQ at
+[https://www.contributor-covenant.org/faq][FAQ]. Translations are available at
+[https://www.contributor-covenant.org/translations][translations].
+
+[homepage]: https://www.contributor-covenant.org
+[v2.1]: https://www.contributor-covenant.org/version/2/1/code_of_conduct.html
+[Mozilla CoC]: https://github.com/mozilla/diversity
+[FAQ]: https://www.contributor-covenant.org/faq
+[translations]: https://www.contributor-covenant.org/translations
diff --git a/CONTRIBUTING.md b/CONTRIBUTING.md
new file mode 100644
index 00000000..dbf459d2
--- /dev/null
+++ b/CONTRIBUTING.md
@@ -0,0 +1,32 @@
+# How to contribute
+So you want to write code and get it landed in the official OMRChecker repository?
+First, fork our repository into your own GitHub account, and create a local clone of it as described in the installation instructions.
+The latter will be used to get new features implemented or bugs fixed.
+
+Once done and you have the code locally on the disk, you can get started. We advise you to not work directly on the master branch,
+but to create a separate branch for each issue you are working on. That way you can easily switch between different work,
+and you can update each one for the latest changes on the upstream master individually.
+
+
+# Writing Code
+For writing the code just follow the [Pep8 Python style](https://peps.python.org/pep-0008/) guide, If there is something unclear about the style, just look at existing code which might help you to understand it better.
+
+Also, try to use commits with [conventional messages](https://www.conventionalcommits.org/en/v1.0.0/#summary).
+
+
+# Code Formatting
+Before committing your code, make sure to run the following command to format your code according to the PEP8 style guide:
+```.sh
+pip install -r requirements.dev.txt && pre-commit install
+```
+
+Run `pre-commit` before committing your changes:
+```.sh
+git add .
+pre-commit run -a
+```
+
+# Where to contribute from
+
+- You can pickup any open [issues](https://github.com/Udayraj123/OMRChecker/issues) to solve.
+- You can also check out the [ideas list](https://github.com/users/Udayraj123/projects/2/views/1)
diff --git a/Contributors.md b/Contributors.md
new file mode 100644
index 00000000..3f95b245
--- /dev/null
+++ b/Contributors.md
@@ -0,0 +1,22 @@
+# Contributors
+
+- [Udayraj123](https://github.com/Udayraj123)
+- [leongwaikay](https://github.com/leongwaikay)
+- [deepakgouda](https://github.com/deepakgouda)
+- [apurva91](https://github.com/apurva91)
+- [sparsh2706](https://github.com/sparsh2706)
+- [namit2saxena](https://github.com/namit2saxena)
+- [Harsh-Kapoorr](https://github.com/Harsh-Kapoorr)
+- [Sandeep-1507](https://github.com/Sandeep-1507)
+- [SpyzzVVarun](https://github.com/SpyzzVVarun)
+- [asc249](https://github.com/asc249)
+- [05Alston](https://github.com/05Alston)
+- [Antibodyy](https://github.com/Antibodyy)
+- [infinity1729](https://github.com/infinity1729)
+- [Rohan-G](https://github.com/Rohan-G)
+- [UjjwalMahar](https://github.com/UjjwalMahar)
+- [Kurtsley](https://github.com/Kurtsley)
+- [gaursagar21](https://github.com/gaursagar21)
+- [aayushibansal2001](https://github.com/aayushibansal2001)
+- [ShamanthVallem](https://github.com/ShamanthVallem)
+- [rudrapsc](https://github.com/rudrapsc)
diff --git a/LICENSE b/LICENSE
index fbbea73a..3942f30c 100644
--- a/LICENSE
+++ b/LICENSE
@@ -1,621 +1,22 @@
- GNU GENERAL PUBLIC LICENSE
- Version 3, 29 June 2007
-
- Copyright (C) 2007 Free Software Foundation, Inc.
- Everyone is permitted to copy and distribute verbatim copies
- of this license document, but changing it is not allowed.
-
- Preamble
-
- The GNU General Public License is a free, copyleft license for
-software and other kinds of works.
-
- The licenses for most software and other practical works are designed
-to take away your freedom to share and change the works. By contrast,
-the GNU General Public License is intended to guarantee your freedom to
-share and change all versions of a program--to make sure it remains free
-software for all its users. We, the Free Software Foundation, use the
-GNU General Public License for most of our software; it applies also to
-any other work released this way by its authors. You can apply it to
-your programs, too.
-
- When we speak of free software, we are referring to freedom, not
-price. Our General Public Licenses are designed to make sure that you
-have the freedom to distribute copies of free software (and charge for
-them if you wish), that you receive source code or can get it if you
-want it, that you can change the software or use pieces of it in new
-free programs, and that you know you can do these things.
-
- To protect your rights, we need to prevent others from denying you
-these rights or asking you to surrender the rights. Therefore, you have
-certain responsibilities if you distribute copies of the software, or if
-you modify it: responsibilities to respect the freedom of others.
-
- For example, if you distribute copies of such a program, whether
-gratis or for a fee, you must pass on to the recipients the same
-freedoms that you received. You must make sure that they, too, receive
-or can get the source code. And you must show them these terms so they
-know their rights.
-
- Developers that use the GNU GPL protect your rights with two steps:
-(1) assert copyright on the software, and (2) offer you this License
-giving you legal permission to copy, distribute and/or modify it.
-
- For the developers' and authors' protection, the GPL clearly explains
-that there is no warranty for this free software. For both users' and
-authors' sake, the GPL requires that modified versions be marked as
-changed, so that their problems will not be attributed erroneously to
-authors of previous versions.
-
- Some devices are designed to deny users access to install or run
-modified versions of the software inside them, although the manufacturer
-can do so. This is fundamentally incompatible with the aim of
-protecting users' freedom to change the software. The systematic
-pattern of such abuse occurs in the area of products for individuals to
-use, which is precisely where it is most unacceptable. Therefore, we
-have designed this version of the GPL to prohibit the practice for those
-products. If such problems arise substantially in other domains, we
-stand ready to extend this provision to those domains in future versions
-of the GPL, as needed to protect the freedom of users.
-
- Finally, every program is threatened constantly by software patents.
-States should not allow patents to restrict development and use of
-software on general-purpose computers, but in those that do, we wish to
-avoid the special danger that patents applied to a free program could
-make it effectively proprietary. To prevent this, the GPL assures that
-patents cannot be used to render the program non-free.
-
- The precise terms and conditions for copying, distribution and
-modification follow.
-
- TERMS AND CONDITIONS
-
- 0. Definitions.
-
- "This License" refers to version 3 of the GNU General Public License.
-
- "Copyright" also means copyright-like laws that apply to other kinds of
-works, such as semiconductor masks.
-
- "The Program" refers to any copyrightable work licensed under this
-License. Each licensee is addressed as "you". "Licensees" and
-"recipients" may be individuals or organizations.
-
- To "modify" a work means to copy from or adapt all or part of the work
-in a fashion requiring copyright permission, other than the making of an
-exact copy. The resulting work is called a "modified version" of the
-earlier work or a work "based on" the earlier work.
-
- A "covered work" means either the unmodified Program or a work based
-on the Program.
-
- To "propagate" a work means to do anything with it that, without
-permission, would make you directly or secondarily liable for
-infringement under applicable copyright law, except executing it on a
-computer or modifying a private copy. Propagation includes copying,
-distribution (with or without modification), making available to the
-public, and in some countries other activities as well.
-
- To "convey" a work means any kind of propagation that enables other
-parties to make or receive copies. Mere interaction with a user through
-a computer network, with no transfer of a copy, is not conveying.
-
- An interactive user interface displays "Appropriate Legal Notices"
-to the extent that it includes a convenient and prominently visible
-feature that (1) displays an appropriate copyright notice, and (2)
-tells the user that there is no warranty for the work (except to the
-extent that warranties are provided), that licensees may convey the
-work under this License, and how to view a copy of this License. If
-the interface presents a list of user commands or options, such as a
-menu, a prominent item in the list meets this criterion.
-
- 1. Source Code.
-
- The "source code" for a work means the preferred form of the work
-for making modifications to it. "Object code" means any non-source
-form of a work.
-
- A "Standard Interface" means an interface that either is an official
-standard defined by a recognized standards body, or, in the case of
-interfaces specified for a particular programming language, one that
-is widely used among developers working in that language.
-
- The "System Libraries" of an executable work include anything, other
-than the work as a whole, that (a) is included in the normal form of
-packaging a Major Component, but which is not part of that Major
-Component, and (b) serves only to enable use of the work with that
-Major Component, or to implement a Standard Interface for which an
-implementation is available to the public in source code form. A
-"Major Component", in this context, means a major essential component
-(kernel, window system, and so on) of the specific operating system
-(if any) on which the executable work runs, or a compiler used to
-produce the work, or an object code interpreter used to run it.
-
- The "Corresponding Source" for a work in object code form means all
-the source code needed to generate, install, and (for an executable
-work) run the object code and to modify the work, including scripts to
-control those activities. However, it does not include the work's
-System Libraries, or general-purpose tools or generally available free
-programs which are used unmodified in performing those activities but
-which are not part of the work. For example, Corresponding Source
-includes interface definition files associated with source files for
-the work, and the source code for shared libraries and dynamically
-linked subprograms that the work is specifically designed to require,
-such as by intimate data communication or control flow between those
-subprograms and other parts of the work.
-
- The Corresponding Source need not include anything that users
-can regenerate automatically from other parts of the Corresponding
-Source.
-
- The Corresponding Source for a work in source code form is that
-same work.
-
- 2. Basic Permissions.
-
- All rights granted under this License are granted for the term of
-copyright on the Program, and are irrevocable provided the stated
-conditions are met. This License explicitly affirms your unlimited
-permission to run the unmodified Program. The output from running a
-covered work is covered by this License only if the output, given its
-content, constitutes a covered work. This License acknowledges your
-rights of fair use or other equivalent, as provided by copyright law.
-
- You may make, run and propagate covered works that you do not
-convey, without conditions so long as your license otherwise remains
-in force. You may convey covered works to others for the sole purpose
-of having them make modifications exclusively for you, or provide you
-with facilities for running those works, provided that you comply with
-the terms of this License in conveying all material for which you do
-not control copyright. Those thus making or running the covered works
-for you must do so exclusively on your behalf, under your direction
-and control, on terms that prohibit them from making any copies of
-your copyrighted material outside their relationship with you.
-
- Conveying under any other circumstances is permitted solely under
-the conditions stated below. Sublicensing is not allowed; section 10
-makes it unnecessary.
-
- 3. Protecting Users' Legal Rights From Anti-Circumvention Law.
-
- No covered work shall be deemed part of an effective technological
-measure under any applicable law fulfilling obligations under article
-11 of the WIPO copyright treaty adopted on 20 December 1996, or
-similar laws prohibiting or restricting circumvention of such
-measures.
-
- When you convey a covered work, you waive any legal power to forbid
-circumvention of technological measures to the extent such circumvention
-is effected by exercising rights under this License with respect to
-the covered work, and you disclaim any intention to limit operation or
-modification of the work as a means of enforcing, against the work's
-users, your or third parties' legal rights to forbid circumvention of
-technological measures.
-
- 4. Conveying Verbatim Copies.
-
- You may convey verbatim copies of the Program's source code as you
-receive it, in any medium, provided that you conspicuously and
-appropriately publish on each copy an appropriate copyright notice;
-keep intact all notices stating that this License and any
-non-permissive terms added in accord with section 7 apply to the code;
-keep intact all notices of the absence of any warranty; and give all
-recipients a copy of this License along with the Program.
-
- You may charge any price or no price for each copy that you convey,
-and you may offer support or warranty protection for a fee.
-
- 5. Conveying Modified Source Versions.
-
- You may convey a work based on the Program, or the modifications to
-produce it from the Program, in the form of source code under the
-terms of section 4, provided that you also meet all of these conditions:
-
- a) The work must carry prominent notices stating that you modified
- it, and giving a relevant date.
-
- b) The work must carry prominent notices stating that it is
- released under this License and any conditions added under section
- 7. This requirement modifies the requirement in section 4 to
- "keep intact all notices".
-
- c) You must license the entire work, as a whole, under this
- License to anyone who comes into possession of a copy. This
- License will therefore apply, along with any applicable section 7
- additional terms, to the whole of the work, and all its parts,
- regardless of how they are packaged. This License gives no
- permission to license the work in any other way, but it does not
- invalidate such permission if you have separately received it.
-
- d) If the work has interactive user interfaces, each must display
- Appropriate Legal Notices; however, if the Program has interactive
- interfaces that do not display Appropriate Legal Notices, your
- work need not make them do so.
-
- A compilation of a covered work with other separate and independent
-works, which are not by their nature extensions of the covered work,
-and which are not combined with it such as to form a larger program,
-in or on a volume of a storage or distribution medium, is called an
-"aggregate" if the compilation and its resulting copyright are not
-used to limit the access or legal rights of the compilation's users
-beyond what the individual works permit. Inclusion of a covered work
-in an aggregate does not cause this License to apply to the other
-parts of the aggregate.
-
- 6. Conveying Non-Source Forms.
-
- You may convey a covered work in object code form under the terms
-of sections 4 and 5, provided that you also convey the
-machine-readable Corresponding Source under the terms of this License,
-in one of these ways:
-
- a) Convey the object code in, or embodied in, a physical product
- (including a physical distribution medium), accompanied by the
- Corresponding Source fixed on a durable physical medium
- customarily used for software interchange.
-
- b) Convey the object code in, or embodied in, a physical product
- (including a physical distribution medium), accompanied by a
- written offer, valid for at least three years and valid for as
- long as you offer spare parts or customer support for that product
- model, to give anyone who possesses the object code either (1) a
- copy of the Corresponding Source for all the software in the
- product that is covered by this License, on a durable physical
- medium customarily used for software interchange, for a price no
- more than your reasonable cost of physically performing this
- conveying of source, or (2) access to copy the
- Corresponding Source from a network server at no charge.
-
- c) Convey individual copies of the object code with a copy of the
- written offer to provide the Corresponding Source. This
- alternative is allowed only occasionally and noncommercially, and
- only if you received the object code with such an offer, in accord
- with subsection 6b.
-
- d) Convey the object code by offering access from a designated
- place (gratis or for a charge), and offer equivalent access to the
- Corresponding Source in the same way through the same place at no
- further charge. You need not require recipients to copy the
- Corresponding Source along with the object code. If the place to
- copy the object code is a network server, the Corresponding Source
- may be on a different server (operated by you or a third party)
- that supports equivalent copying facilities, provided you maintain
- clear directions next to the object code saying where to find the
- Corresponding Source. Regardless of what server hosts the
- Corresponding Source, you remain obligated to ensure that it is
- available for as long as needed to satisfy these requirements.
-
- e) Convey the object code using peer-to-peer transmission, provided
- you inform other peers where the object code and Corresponding
- Source of the work are being offered to the general public at no
- charge under subsection 6d.
-
- A separable portion of the object code, whose source code is excluded
-from the Corresponding Source as a System Library, need not be
-included in conveying the object code work.
-
- A "User Product" is either (1) a "consumer product", which means any
-tangible personal property which is normally used for personal, family,
-or household purposes, or (2) anything designed or sold for incorporation
-into a dwelling. In determining whether a product is a consumer product,
-doubtful cases shall be resolved in favor of coverage. For a particular
-product received by a particular user, "normally used" refers to a
-typical or common use of that class of product, regardless of the status
-of the particular user or of the way in which the particular user
-actually uses, or expects or is expected to use, the product. A product
-is a consumer product regardless of whether the product has substantial
-commercial, industrial or non-consumer uses, unless such uses represent
-the only significant mode of use of the product.
-
- "Installation Information" for a User Product means any methods,
-procedures, authorization keys, or other information required to install
-and execute modified versions of a covered work in that User Product from
-a modified version of its Corresponding Source. The information must
-suffice to ensure that the continued functioning of the modified object
-code is in no case prevented or interfered with solely because
-modification has been made.
-
- If you convey an object code work under this section in, or with, or
-specifically for use in, a User Product, and the conveying occurs as
-part of a transaction in which the right of possession and use of the
-User Product is transferred to the recipient in perpetuity or for a
-fixed term (regardless of how the transaction is characterized), the
-Corresponding Source conveyed under this section must be accompanied
-by the Installation Information. But this requirement does not apply
-if neither you nor any third party retains the ability to install
-modified object code on the User Product (for example, the work has
-been installed in ROM).
-
- The requirement to provide Installation Information does not include a
-requirement to continue to provide support service, warranty, or updates
-for a work that has been modified or installed by the recipient, or for
-the User Product in which it has been modified or installed. Access to a
-network may be denied when the modification itself materially and
-adversely affects the operation of the network or violates the rules and
-protocols for communication across the network.
-
- Corresponding Source conveyed, and Installation Information provided,
-in accord with this section must be in a format that is publicly
-documented (and with an implementation available to the public in
-source code form), and must require no special password or key for
-unpacking, reading or copying.
-
- 7. Additional Terms.
-
- "Additional permissions" are terms that supplement the terms of this
-License by making exceptions from one or more of its conditions.
-Additional permissions that are applicable to the entire Program shall
-be treated as though they were included in this License, to the extent
-that they are valid under applicable law. If additional permissions
-apply only to part of the Program, that part may be used separately
-under those permissions, but the entire Program remains governed by
-this License without regard to the additional permissions.
-
- When you convey a copy of a covered work, you may at your option
-remove any additional permissions from that copy, or from any part of
-it. (Additional permissions may be written to require their own
-removal in certain cases when you modify the work.) You may place
-additional permissions on material, added by you to a covered work,
-for which you have or can give appropriate copyright permission.
-
- Notwithstanding any other provision of this License, for material you
-add to a covered work, you may (if authorized by the copyright holders of
-that material) supplement the terms of this License with terms:
-
- a) Disclaiming warranty or limiting liability differently from the
- terms of sections 15 and 16 of this License; or
-
- b) Requiring preservation of specified reasonable legal notices or
- author attributions in that material or in the Appropriate Legal
- Notices displayed by works containing it; or
-
- c) Prohibiting misrepresentation of the origin of that material, or
- requiring that modified versions of such material be marked in
- reasonable ways as different from the original version; or
-
- d) Limiting the use for publicity purposes of names of licensors or
- authors of the material; or
-
- e) Declining to grant rights under trademark law for use of some
- trade names, trademarks, or service marks; or
-
- f) Requiring indemnification of licensors and authors of that
- material by anyone who conveys the material (or modified versions of
- it) with contractual assumptions of liability to the recipient, for
- any liability that these contractual assumptions directly impose on
- those licensors and authors.
-
- All other non-permissive additional terms are considered "further
-restrictions" within the meaning of section 10. If the Program as you
-received it, or any part of it, contains a notice stating that it is
-governed by this License along with a term that is a further
-restriction, you may remove that term. If a license document contains
-a further restriction but permits relicensing or conveying under this
-License, you may add to a covered work material governed by the terms
-of that license document, provided that the further restriction does
-not survive such relicensing or conveying.
-
- If you add terms to a covered work in accord with this section, you
-must place, in the relevant source files, a statement of the
-additional terms that apply to those files, or a notice indicating
-where to find the applicable terms.
-
- Additional terms, permissive or non-permissive, may be stated in the
-form of a separately written license, or stated as exceptions;
-the above requirements apply either way.
-
- 8. Termination.
-
- You may not propagate or modify a covered work except as expressly
-provided under this License. Any attempt otherwise to propagate or
-modify it is void, and will automatically terminate your rights under
-this License (including any patent licenses granted under the third
-paragraph of section 11).
-
- However, if you cease all violation of this License, then your
-license from a particular copyright holder is reinstated (a)
-provisionally, unless and until the copyright holder explicitly and
-finally terminates your license, and (b) permanently, if the copyright
-holder fails to notify you of the violation by some reasonable means
-prior to 60 days after the cessation.
-
- Moreover, your license from a particular copyright holder is
-reinstated permanently if the copyright holder notifies you of the
-violation by some reasonable means, this is the first time you have
-received notice of violation of this License (for any work) from that
-copyright holder, and you cure the violation prior to 30 days after
-your receipt of the notice.
-
- Termination of your rights under this section does not terminate the
-licenses of parties who have received copies or rights from you under
-this License. If your rights have been terminated and not permanently
-reinstated, you do not qualify to receive new licenses for the same
-material under section 10.
-
- 9. Acceptance Not Required for Having Copies.
-
- You are not required to accept this License in order to receive or
-run a copy of the Program. Ancillary propagation of a covered work
-occurring solely as a consequence of using peer-to-peer transmission
-to receive a copy likewise does not require acceptance. However,
-nothing other than this License grants you permission to propagate or
-modify any covered work. These actions infringe copyright if you do
-not accept this License. Therefore, by modifying or propagating a
-covered work, you indicate your acceptance of this License to do so.
-
- 10. Automatic Licensing of Downstream Recipients.
-
- Each time you convey a covered work, the recipient automatically
-receives a license from the original licensors, to run, modify and
-propagate that work, subject to this License. You are not responsible
-for enforcing compliance by third parties with this License.
-
- An "entity transaction" is a transaction transferring control of an
-organization, or substantially all assets of one, or subdividing an
-organization, or merging organizations. If propagation of a covered
-work results from an entity transaction, each party to that
-transaction who receives a copy of the work also receives whatever
-licenses to the work the party's predecessor in interest had or could
-give under the previous paragraph, plus a right to possession of the
-Corresponding Source of the work from the predecessor in interest, if
-the predecessor has it or can get it with reasonable efforts.
-
- You may not impose any further restrictions on the exercise of the
-rights granted or affirmed under this License. For example, you may
-not impose a license fee, royalty, or other charge for exercise of
-rights granted under this License, and you may not initiate litigation
-(including a cross-claim or counterclaim in a lawsuit) alleging that
-any patent claim is infringed by making, using, selling, offering for
-sale, or importing the Program or any portion of it.
-
- 11. Patents.
-
- A "contributor" is a copyright holder who authorizes use under this
-License of the Program or a work on which the Program is based. The
-work thus licensed is called the contributor's "contributor version".
-
- A contributor's "essential patent claims" are all patent claims
-owned or controlled by the contributor, whether already acquired or
-hereafter acquired, that would be infringed by some manner, permitted
-by this License, of making, using, or selling its contributor version,
-but do not include claims that would be infringed only as a
-consequence of further modification of the contributor version. For
-purposes of this definition, "control" includes the right to grant
-patent sublicenses in a manner consistent with the requirements of
-this License.
-
- Each contributor grants you a non-exclusive, worldwide, royalty-free
-patent license under the contributor's essential patent claims, to
-make, use, sell, offer for sale, import and otherwise run, modify and
-propagate the contents of its contributor version.
-
- In the following three paragraphs, a "patent license" is any express
-agreement or commitment, however denominated, not to enforce a patent
-(such as an express permission to practice a patent or covenant not to
-sue for patent infringement). To "grant" such a patent license to a
-party means to make such an agreement or commitment not to enforce a
-patent against the party.
-
- If you convey a covered work, knowingly relying on a patent license,
-and the Corresponding Source of the work is not available for anyone
-to copy, free of charge and under the terms of this License, through a
-publicly available network server or other readily accessible means,
-then you must either (1) cause the Corresponding Source to be so
-available, or (2) arrange to deprive yourself of the benefit of the
-patent license for this particular work, or (3) arrange, in a manner
-consistent with the requirements of this License, to extend the patent
-license to downstream recipients. "Knowingly relying" means you have
-actual knowledge that, but for the patent license, your conveying the
-covered work in a country, or your recipient's use of the covered work
-in a country, would infringe one or more identifiable patents in that
-country that you have reason to believe are valid.
-
- If, pursuant to or in connection with a single transaction or
-arrangement, you convey, or propagate by procuring conveyance of, a
-covered work, and grant a patent license to some of the parties
-receiving the covered work authorizing them to use, propagate, modify
-or convey a specific copy of the covered work, then the patent license
-you grant is automatically extended to all recipients of the covered
-work and works based on it.
-
- A patent license is "discriminatory" if it does not include within
-the scope of its coverage, prohibits the exercise of, or is
-conditioned on the non-exercise of one or more of the rights that are
-specifically granted under this License. You may not convey a covered
-work if you are a party to an arrangement with a third party that is
-in the business of distributing software, under which you make payment
-to the third party based on the extent of your activity of conveying
-the work, and under which the third party grants, to any of the
-parties who would receive the covered work from you, a discriminatory
-patent license (a) in connection with copies of the covered work
-conveyed by you (or copies made from those copies), or (b) primarily
-for and in connection with specific products or compilations that
-contain the covered work, unless you entered into that arrangement,
-or that patent license was granted, prior to 28 March 2007.
-
- Nothing in this License shall be construed as excluding or limiting
-any implied license or other defenses to infringement that may
-otherwise be available to you under applicable patent law.
-
- 12. No Surrender of Others' Freedom.
-
- If conditions are imposed on you (whether by court order, agreement or
-otherwise) that contradict the conditions of this License, they do not
-excuse you from the conditions of this License. If you cannot convey a
-covered work so as to satisfy simultaneously your obligations under this
-License and any other pertinent obligations, then as a consequence you may
-not convey it at all. For example, if you agree to terms that obligate you
-to collect a royalty for further conveying from those to whom you convey
-the Program, the only way you could satisfy both those terms and this
-License would be to refrain entirely from conveying the Program.
-
- 13. Use with the GNU Affero General Public License.
-
- Notwithstanding any other provision of this License, you have
-permission to link or combine any covered work with a work licensed
-under version 3 of the GNU Affero General Public License into a single
-combined work, and to convey the resulting work. The terms of this
-License will continue to apply to the part which is the covered work,
-but the special requirements of the GNU Affero General Public License,
-section 13, concerning interaction through a network will apply to the
-combination as such.
-
- 14. Revised Versions of this License.
-
- The Free Software Foundation may publish revised and/or new versions of
-the GNU General Public License from time to time. Such new versions will
-be similar in spirit to the present version, but may differ in detail to
-address new problems or concerns.
-
- Each version is given a distinguishing version number. If the
-Program specifies that a certain numbered version of the GNU General
-Public License "or any later version" applies to it, you have the
-option of following the terms and conditions either of that numbered
-version or of any later version published by the Free Software
-Foundation. If the Program does not specify a version number of the
-GNU General Public License, you may choose any version ever published
-by the Free Software Foundation.
-
- If the Program specifies that a proxy can decide which future
-versions of the GNU General Public License can be used, that proxy's
-public statement of acceptance of a version permanently authorizes you
-to choose that version for the Program.
-
- Later license versions may give you additional or different
-permissions. However, no additional obligations are imposed on any
-author or copyright holder as a result of your choosing to follow a
-later version.
-
- 15. Disclaimer of Warranty.
-
- THERE IS NO WARRANTY FOR THE PROGRAM, TO THE EXTENT PERMITTED BY
-APPLICABLE LAW. EXCEPT WHEN OTHERWISE STATED IN WRITING THE COPYRIGHT
-HOLDERS AND/OR OTHER PARTIES PROVIDE THE PROGRAM "AS IS" WITHOUT WARRANTY
-OF ANY KIND, EITHER EXPRESSED OR IMPLIED, INCLUDING, BUT NOT LIMITED TO,
-THE IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR
-PURPOSE. THE ENTIRE RISK AS TO THE QUALITY AND PERFORMANCE OF THE PROGRAM
-IS WITH YOU. SHOULD THE PROGRAM PROVE DEFECTIVE, YOU ASSUME THE COST OF
-ALL NECESSARY SERVICING, REPAIR OR CORRECTION.
-
- 16. Limitation of Liability.
-
- IN NO EVENT UNLESS REQUIRED BY APPLICABLE LAW OR AGREED TO IN WRITING
-WILL ANY COPYRIGHT HOLDER, OR ANY OTHER PARTY WHO MODIFIES AND/OR CONVEYS
-THE PROGRAM AS PERMITTED ABOVE, BE LIABLE TO YOU FOR DAMAGES, INCLUDING ANY
-GENERAL, SPECIAL, INCIDENTAL OR CONSEQUENTIAL DAMAGES ARISING OUT OF THE
-USE OR INABILITY TO USE THE PROGRAM (INCLUDING BUT NOT LIMITED TO LOSS OF
-DATA OR DATA BEING RENDERED INACCURATE OR LOSSES SUSTAINED BY YOU OR THIRD
-PARTIES OR A FAILURE OF THE PROGRAM TO OPERATE WITH ANY OTHER PROGRAMS),
-EVEN IF SUCH HOLDER OR OTHER PARTY HAS BEEN ADVISED OF THE POSSIBILITY OF
-SUCH DAMAGES.
-
- 17. Interpretation of Sections 15 and 16.
-
- If the disclaimer of warranty and limitation of liability provided
-above cannot be given local legal effect according to their terms,
-reviewing courts shall apply local law that most closely approximates
-an absolute waiver of all civil liability in connection with the
-Program, unless a warranty or assumption of liability accompanies a
-copy of the Program in return for a fee.
-
- END OF TERMS AND CONDITIONS
\ No newline at end of file
+MIT License
+
+Copyright (c) 2024-present Udayraj Deshmukh and other contributors
+
+Permission is hereby granted, free of charge, to any person obtaining
+a copy of this software and associated documentation files (the
+"Software"), to deal in the Software without restriction, including
+without limitation the rights to use, copy, modify, merge, publish,
+distribute, sublicense, and/or sell copies of the Software, and to
+permit persons to whom the Software is furnished to do so, subject to
+the following conditions:
+
+The above copyright notice and this permission notice shall be
+included in all copies or substantial portions of the Software.
+
+THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND,
+EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF
+MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND
+NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE
+LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION
+OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION
+WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE.
diff --git a/README.md b/README.md
index 8afb795e..279aaca8 100644
--- a/README.md
+++ b/README.md
@@ -1,60 +1,83 @@
# OMR Checker
-Grade exams fast and accurately using a scanner 🖨 or your phone 🤳.
-[](http://hits.dwyl.io/udayraj123/OMRchecker)
-[](https://github.com/Udayraj123/OMRChecker/wiki/TODOs)
+Read OMR sheets fast and accurately using a scanner 🖨 or your phone 🤳.
+
+## What is OMR?
+
+OMR stands for Optical Mark Recognition, used to detect and interpret human-marked data on documents. OMR refers to the process of reading and evaluating OMR sheets, commonly used in exams, surveys, and other forms.
+
+#### **Quick Links**
+
+- [Installation](#getting-started)
+- [User Guide](https://github.com/Udayraj123/OMRChecker/wiki)
+- [Contributor Guide](https://github.com/Udayraj123/OMRChecker/blob/master/CONTRIBUTING.md)
+- [Project Ideas List](https://github.com/users/Udayraj123/projects/2/views/1)
+
+
+
+[](https://github.com/Udayraj123/OMRChecker/pull/new/master)
[](https://github.com/Udayraj123/OMRChecker/pulls?q=is%3Aclosed)
[](https://GitHub.com/Udayraj123/OMRChecker/issues?q=is%3Aissue+is%3Aclosed)
-[](https://GitHub.com/Udayraj123/OMRChecker/graphs/contributors/)
+[](https://github.com/Udayraj123/OMRChecker/issues/5)
+
+
[](https://GitHub.com/Udayraj123/OMRChecker/stargazers/)
-[](https://github.com/Udayraj123/OMRChecker/pull/new/master)
+[](https://hits.seeyoufarm.com)
[](https://discord.gg/qFv2Vqf)
-[](https://github.com/Udayraj123/OMRChecker/issues/5)
-
-
-#### **TLDR;** Jump to [Getting Started](#getting-started).
+
## 🎯 Features
-A full-fledged OMR checking software that can read and evaluate OMR sheets scanned at any angle and having any color. Support is also provided for a customisable marking scheme with section-wise marking, bonus questions, etc.
+A full-fledged OMR checking software that can read and evaluate OMR sheets scanned at any angle and having any color.
+
+| Specs ![]() |  
|   ![]() |
+| :--------------------- | :--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
+| 💯 **Accurate** | Currently nearly 100% accurate on good quality document scans; and about 90% accurate on mobile images. |
+| 💪🏿 **Robust** | Supports low resolution, xeroxed sheets. See [**Robustness**](https://github.com/Udayraj123/OMRChecker/wiki/Robustness) for more. |
+| ⏩ **Fast** | Current processing speed without any optimization is 200 OMRs/minute. |
+| ✅ **Customizable** | [Easily apply](https://github.com/Udayraj123/OMRChecker/wiki/User-Guide) to custom OMR layouts, surveys, etc. |
+| 📊 **Visually Rich** | [Get insights](https://github.com/Udayraj123/OMRChecker/wiki/Rich-Visuals) to configure and debug easily. |
+| 🎈 **Lightweight** | Very minimal core code size. |
+| 🏫 **Large Scale** | Tested on a large scale at [Technothlon](https://en.wikipedia.org/wiki/Technothlon). |
+| 👩🏿💻 **Dev Friendly** | [Pylinted](http://pylint.pycqa.org/) and [Black formatted](https://github.com/psf/black) code. Also has a [developer community](https://discord.gg/qFv2Vqf) on discord. |
-| Specs |    |
-|:----------------:|-----------------------------------------------------------------------------------------------------------------------------------|
-| 💯 **Accurate** | Currently nearly 100% accurate on good quality document scans; and about 90% accurate on mobile images. |
-| 💪🏿 **Robust** | Supports low resolution, xeroxed sheets. See [**Robustness**](https://github.com/Udayraj123/OMRChecker/wiki/Robustness) for more. |
-| ⏩ **Fast** | Current processing speed without any optimization is 200 OMRs/minute. |
-| ✅ **Extensible** | [**Easily apply**](https://github.com/Udayraj123/OMRChecker/wiki/User-Guide) to different OMR layouts, surveys, etc. |
-| 📊 **Visually Rich Outputs** | [Get insights](https://github.com/Udayraj123/OMRChecker/wiki/Rich-Visuals) to configure and debug easily. |
-| 🎈 **Extremely lightweight** | Core code size is **less than 500 KB**(Samples excluded). |
-| 🏫 **Large Scale** | Used on tens of thousands of OMRs at [Technothlon](https://www.facebook.com/technothlon.techniche). |
-| 👩🏿💻 **Dev Friendly** | [**Well documented**](https://github.com/Udayraj123/OMRChecker/wiki/) repository based on python and openCV with [an active discussion group](https://discord.gg/qFv2Vqf). |
+Note: For solving interesting challenges, developers can check out [**TODOs**](https://github.com/Udayraj123/OMRChecker/wiki/TODOs).
-Note: For solving live challenges, developers can checkout [**TODOs**](https://github.com/Udayraj123/OMRChecker/wiki/TODOs).
-See all details in [Project Wiki](https://github.com/Udayraj123/OMRChecker/wiki/).
+See the complete guide and details at [Project Wiki](https://github.com/Udayraj123/OMRChecker/wiki/).
+
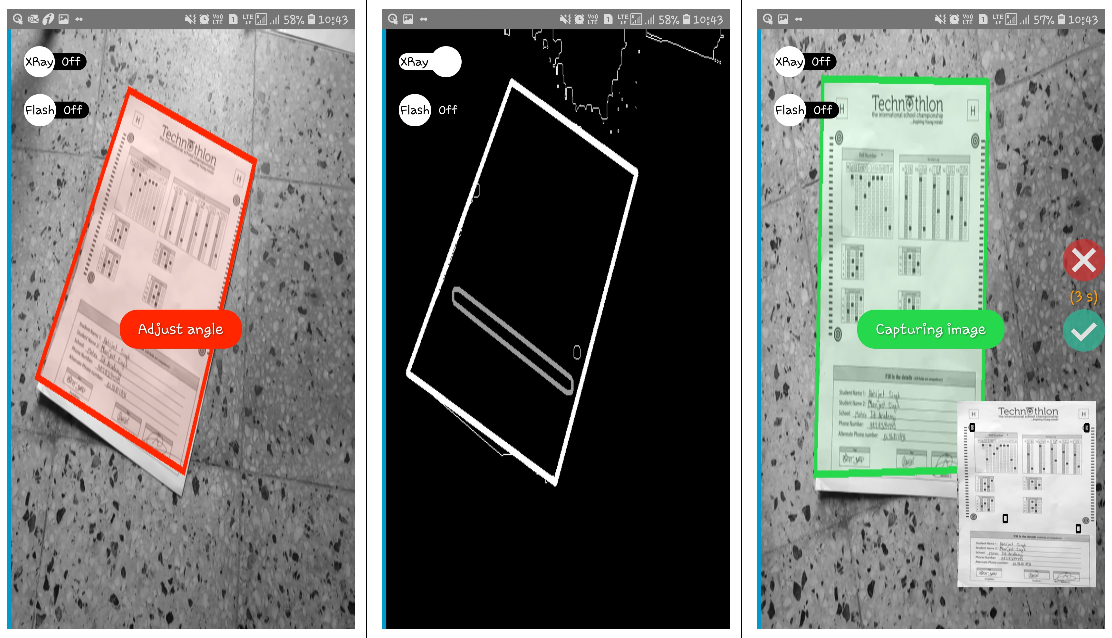
## 💡 What can OMRChecker do for me?
-Once you configure the OMR layout, just throw images of the sheets at the software; and you'll get back the graded responses in an excel sheet!
+
+Once you configure the OMR layout, just throw images of the sheets at the software; and you'll get back the marked responses in an excel sheet!
Images can be taken from various angles as shown below-
+
|
+| :--------------------- | :--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
+| 💯 **Accurate** | Currently nearly 100% accurate on good quality document scans; and about 90% accurate on mobile images. |
+| 💪🏿 **Robust** | Supports low resolution, xeroxed sheets. See [**Robustness**](https://github.com/Udayraj123/OMRChecker/wiki/Robustness) for more. |
+| ⏩ **Fast** | Current processing speed without any optimization is 200 OMRs/minute. |
+| ✅ **Customizable** | [Easily apply](https://github.com/Udayraj123/OMRChecker/wiki/User-Guide) to custom OMR layouts, surveys, etc. |
+| 📊 **Visually Rich** | [Get insights](https://github.com/Udayraj123/OMRChecker/wiki/Rich-Visuals) to configure and debug easily. |
+| 🎈 **Lightweight** | Very minimal core code size. |
+| 🏫 **Large Scale** | Tested on a large scale at [Technothlon](https://en.wikipedia.org/wiki/Technothlon). |
+| 👩🏿💻 **Dev Friendly** | [Pylinted](http://pylint.pycqa.org/) and [Black formatted](https://github.com/psf/black) code. Also has a [developer community](https://discord.gg/qFv2Vqf) on discord. |
-| Specs |    |
-|:----------------:|-----------------------------------------------------------------------------------------------------------------------------------|
-| 💯 **Accurate** | Currently nearly 100% accurate on good quality document scans; and about 90% accurate on mobile images. |
-| 💪🏿 **Robust** | Supports low resolution, xeroxed sheets. See [**Robustness**](https://github.com/Udayraj123/OMRChecker/wiki/Robustness) for more. |
-| ⏩ **Fast** | Current processing speed without any optimization is 200 OMRs/minute. |
-| ✅ **Extensible** | [**Easily apply**](https://github.com/Udayraj123/OMRChecker/wiki/User-Guide) to different OMR layouts, surveys, etc. |
-| 📊 **Visually Rich Outputs** | [Get insights](https://github.com/Udayraj123/OMRChecker/wiki/Rich-Visuals) to configure and debug easily. |
-| 🎈 **Extremely lightweight** | Core code size is **less than 500 KB**(Samples excluded). |
-| 🏫 **Large Scale** | Used on tens of thousands of OMRs at [Technothlon](https://www.facebook.com/technothlon.techniche). |
-| 👩🏿💻 **Dev Friendly** | [**Well documented**](https://github.com/Udayraj123/OMRChecker/wiki/) repository based on python and openCV with [an active discussion group](https://discord.gg/qFv2Vqf). |
+Note: For solving interesting challenges, developers can check out [**TODOs**](https://github.com/Udayraj123/OMRChecker/wiki/TODOs).
-Note: For solving live challenges, developers can checkout [**TODOs**](https://github.com/Udayraj123/OMRChecker/wiki/TODOs).
-See all details in [Project Wiki](https://github.com/Udayraj123/OMRChecker/wiki/).
+See the complete guide and details at [Project Wiki](https://github.com/Udayraj123/OMRChecker/wiki/).
+
## 💡 What can OMRChecker do for me?
-Once you configure the OMR layout, just throw images of the sheets at the software; and you'll get back the graded responses in an excel sheet!
+
+Once you configure the OMR layout, just throw images of the sheets at the software; and you'll get back the marked responses in an excel sheet!
Images can be taken from various angles as shown below-
+

-### Code in action on images taken by scanner:
+### Code in action on images taken by scanner:
+
-### Code in action on images taken by a mobile phone:
+### Code in action on images taken by a mobile phone:
+
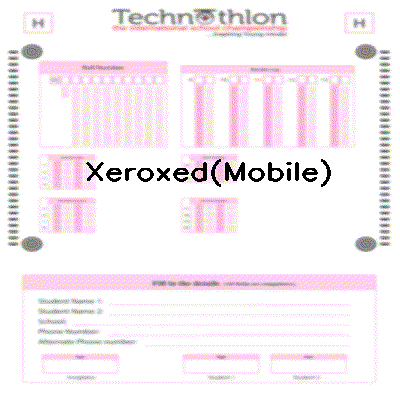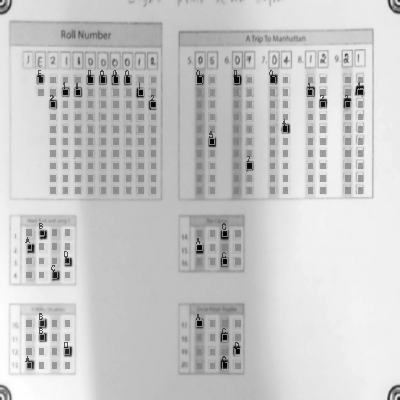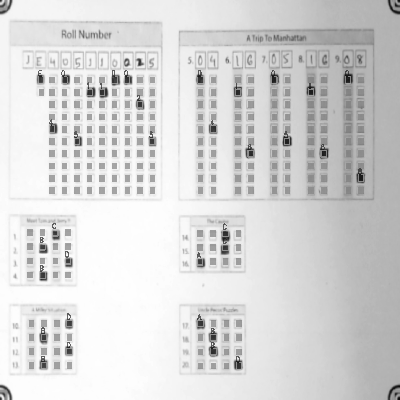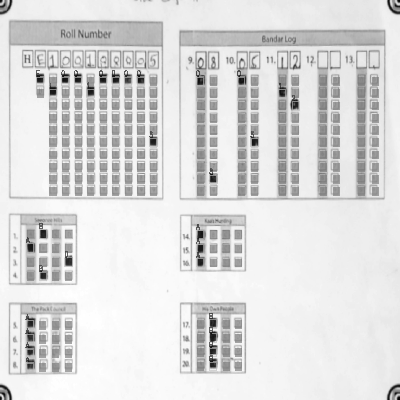
-See step by step processing of any OMR sheet:
+## Visuals
+
+### Processing steps
+
+See step-by-step processing of any OMR sheet:
+
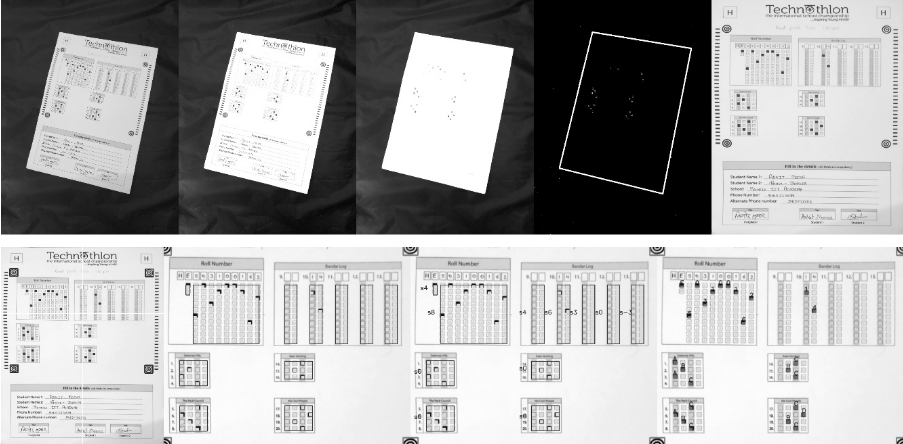 @@ -63,7 +86,9 @@ See step by step processing of any OMR sheet:
*Note: This image is generated by the code itself!*
@@ -63,7 +86,9 @@ See step by step processing of any OMR sheet:
*Note: This image is generated by the code itself!*
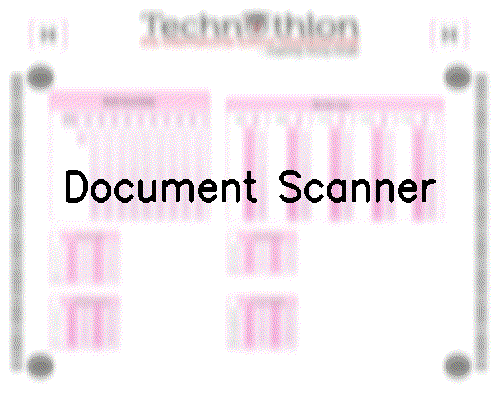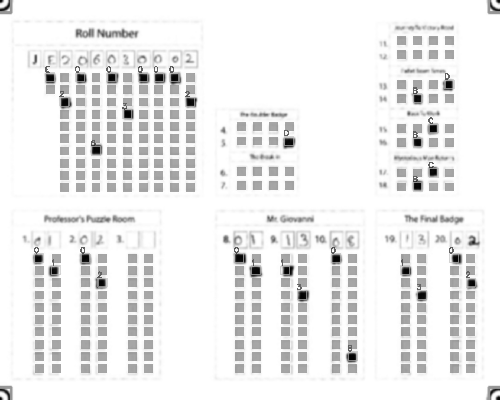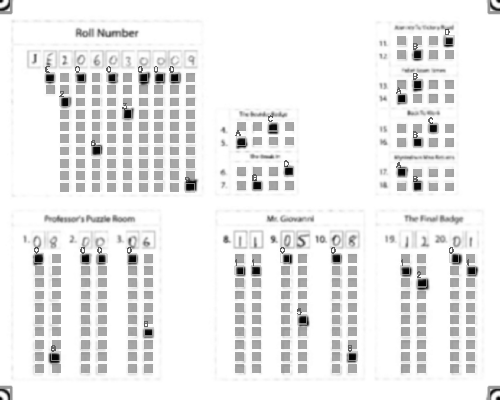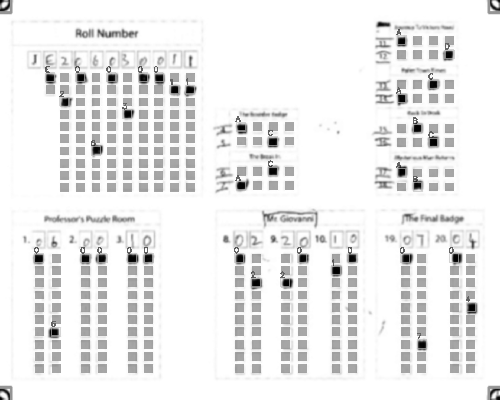
-Output: A CSV sheet containing the detected responses and evaluated scores:
+### Output
+
+Get a CSV sheet containing the detected responses and evaluated scores:
@@ -71,29 +96,65 @@ Output: A CSV sheet containing the detected responses and evaluated scores:
-#### There are many visuals in the wiki. [Check them out!](https://github.com/Udayraj123/OMRChecker/wiki/Rich-Visuals)
+We now support [colored outputs](https://github.com/Udayraj123/OMRChecker/wiki/%5Bv2%5D-About-Evaluation) as well. Here's a sample output on another image -
+
+
+  +
+
+
+
+
+#### There are many more visuals in the wiki. Check them out [here!](https://github.com/Udayraj123/OMRChecker/wiki/Rich-Visuals)
## Getting started
+

-### Operating System
-Although windows is supported, **Linux** is recommended for a bug-free experience.
+**Operating system:** OSX or Linux is recommended although Windows is also supported.
-### 1. Install dependencies
- 
+### 1. Install global dependencies
+
+ 
+
+To check if python3 and pip is already installed:
+
+```bash
+python3 --version
+python3 -m pip --version
+```
+
+
+ Install Python3
+
+To install python3 follow instructions [here](https://www.python.org/downloads/)
+
+To install pip - follow instructions [here](https://pip.pypa.io/en/stable/installation/)
+
+
+
+Install OpenCV
+
+**Any installation method is fine.**
+
+Recommended:
-_Note: To get a copy button for below commands, use [CodeCopy Chrome](https://chrome.google.com/webstore/detail/codecopy/fkbfebkcoelajmhanocgppanfoojcdmg) | [CodeCopy Firefox](https://addons.mozilla.org/en-US/firefox/addon/codecopy/)._
```bash
python3 -m pip install --user --upgrade pip
python3 -m pip install --user opencv-python
python3 -m pip install --user opencv-contrib-python
```
+
More details on pip install openCV [here](https://www.pyimagesearch.com/2018/09/19/pip-install-opencv/).
-> **Note:** On a fresh computer some of the libraries may get missing in above pip install.
+
+
+
+
+Extra steps(for Linux users only)
+
+Installing missing libraries(if any):
+
+On a fresh computer, some of the libraries may get missing in event after a successful pip install. Install them using following commands[(ref)](https://www.pyimagesearch.com/2018/05/28/ubuntu-18-04-how-to-install-opencv/):
-Install them using the [following commands](https://www.pyimagesearch.com/2018/05/28/ubuntu-18-04-how-to-install-opencv/):
-Windows users may skip this step.
```bash
sudo apt-get install -y build-essential cmake unzip pkg-config
sudo apt-get install -y libjpeg-dev libpng-dev libtiff-dev
@@ -101,126 +162,199 @@ sudo apt-get install -y libavcodec-dev libavformat-dev libswscale-dev libv4l-dev
sudo apt-get install -y libatlas-base-dev gfortran
```
-### 2. Clone the repo
+
+
+### 2. Install project dependencies
+
+Clone the repo
+
```bash
-# Shallow clone - takes latest code with minimal size
-git clone https://github.com/Udayraj123/OMRChecker --depth=1
+git clone https://github.com/Udayraj123/OMRChecker
+cd OMRChecker/
```
-Note: Contributors should take a full clone(without the --depth flag).
-#### Install other requirements
-   
+Install pip requirements
```bash
-cd OMRChecker/
python3 -m pip install --user -r requirements.txt
```
-> **Note:** If you face a distutils error, use `--ignore-installed` flag in above command.
-
-### 3. Run the code
+_**Note:** If you face a distutils error in pip, use `--ignore-installed` flag in above command._
-1. Put your data in inputs folder. You can copy sample data as shown below:
- ```bash
- # Note: you may remove previous inputs if any with `mv inputs/* ~/.trash`
- cp -r ./samples/sample1 inputs/
- ```
- _Note: Change the number N in sampleN to see more examples_
-2. Run OMRChecker:
- **` python3 main.py `**
+
-These samples demonstrate different ways OMRChecker can be used.
+### 3. Run the code
-#### Running it on your own OMR Sheets
+1. First copy and examine the sample data to know how to structure your inputs:
+ ```bash
+ cp -r ./samples/sample1 inputs/
+ # Note: you may remove previous inputs (if any) with `mv inputs/* ~/.trash`
+ # Change the number N in sampleN to see more examples
+ ```
+2. Run OMRChecker:
+ ```bash
+ python3 main.py
+ ```
+
+Alternatively you can also use `python3 main.py -i ./samples/sample1`.
+
+Each example in the samples folder demonstrates different ways in which OMRChecker can be used.
+
+### Common Issues
+
+
+
+ 1. [Windows] ERROR: Could not open requirements file
+
+Command: python3 -m pip install --user -r requirements.txt
+
+ Link to Solution: #54
+
+
+
+2. [Linux] ERROR: No module named pip
+
+Command: python3 -m pip install --user --upgrade pip
+
+ Link to Solution: #70
+
+
+## OMRChecker for custom OMR Sheets
+
+1. First, [create your own template.json](https://github.com/Udayraj123/OMRChecker/wiki/User-Guide).
+2. Configure the tuning parameters.
+3. Run OMRChecker with appropriate arguments (See full usage).
+
+
+## Full Usage
-1. First [create your own template.json](https://github.com/Udayraj123/OMRChecker/wiki/User-Guide).
-2. Open `globals.py` and check the tuning parameters.
-
-3. Run OMRChecker with appropriate arguments.
- #### Full Usage
- ```
- python3 main.py [--setLayout] [--noCropping] [--autoAlign] [--inputDir dir1] [--outputDir dir1] [--template path/to/template.json]
- ```
- Explanation for the arguments:
+```
+python3 main.py [--setLayout] [--inputDir dir1] [--outputDir dir1]
+```
- `--setLayout`: Set up OMR template layout - modify your json file and run again until the template is set.
+Explanation for the arguments:
- `--autoAlign`: (experimental) Enables automatic template alignment - use if the scans show slight misalignments.
+`--setLayout`: Set up OMR template layout - modify your json file and run again until the template is set.
- `--noCropping`: Disables page contour detection - used when page boundary is not visible e.g. document scanner.
+`--inputDir`: Specify an input directory.
- `--inputDir`: Specify an input directory.
+`--outputDir`: Specify an output directory.
- `--outputDir`: Specify an output directory.
+
+
+ Deprecation logs
+
- `--template`: Specify a default template if no template file in input directories.
+- The old `--noCropping` flag has been replaced with the 'CropPage' plugin in "preProcessors" of the template.json(see [samples](https://github.com/Udayraj123/OMRChecker/tree/master/samples)).
+- The `--autoAlign` flag is deprecated due to low performance on a generic OMR sheet
+- The `--template` flag is deprecated and instead it's recommended to keep the template file at the parent folder containing folders of different images
+
-## 💡 Why is this software free?
+## FAQ
+
+
+
+Why is this software free?
+
-The idea for this project began at Technothlon, which is a non-profit international school championship. After seeing it work fabulously at such a large scale, we decided to share this simple and powerful tool with the world to perhaps help revamp OMR checking processes and help greatly reduce the tediousness of the work involved.
+This project was born out of a student-led organization called as [Technothlon](https://technothlon.techniche.org.in). It is a logic-based international school championship organized by students of IIT Guwahati. Being a non-profit organization, and after seeing it work fabulously at such a large scale we decided to share this tool with the world. The OMR checking processes still involves so much tediousness which we aim to reduce dramatically.
-And we believe in the power of open source! Currently, OMRChecker is in its initial stage where only developers can use it. We hope to see it become more user-friendly and even more robust with exposure to different inputs from you!
+We believe in the power of open source! Currently, OMRChecker is in an intermediate stage where only developers can use it. We hope to see it become more user-friendly as well as robust from exposure to different inputs from you all!
[](https://github.com/ellerbrock/open-source-badges/)
-### Can I use this code in my work?
+
+
+
+
+Can I use this code in my (public) work?
+
+
+OMRChecker can be forked and modified. You are encouraged to play with it and we would love to see your own projects in action!
+
+It is published under the [MIT license](https://github.com/Udayraj123/OMRChecker/blob/master/LICENSE).
+
+
+
+
+
+What are the ways to contribute?
+
+
+
+
+- Join the developer community on [Discord](https://discord.gg/qFv2Vqf) to fix [issues](https://github.com/Udayraj123/OMRChecker/issues) with OMRChecker.
+
+- If this project saved you large costs on OMR Software licenses, or saved efforts to make one. Consider donating an amount of your choice(donate section).
-OMRChecker can be forked and modified. **You are encouraged to play with it and we would love to see your own projects in action!** The only requirement is **disclose usage** of this software in your code. It is published under the [**GPLv3 license**](https://github.com/Udayraj123/OMRChecker/blob/master/LICENSE)
+
+
+
-## Credits
-_A Huge thanks to :_
-_The creative master **Adrian Rosebrock** for his blog :_ https://pyimagesearch.com
+
-_The legendary **Harrison** aka sentdex for his [video tutorials](https://www.youtube.com/watch?v=Z78zbnLlPUA&list=PLQVvvaa0QuDdttJXlLtAJxJetJcqmqlQq)._
+## Credits
-_And the james bond of computer vision **Satya Mallic** for his blog:_ https://www.learnopencv.com
+_A Huge thanks to:_
+_**Adrian Rosebrock** for his exemplary blog:_ https://pyimagesearch.com
-_And many other amazing people over the globe without whom this project would never have completed. Thank you!_
+_**Harrison Kinsley** aka sentdex for his [video tutorials](https://www.youtube.com/watch?v=Z78zbnLlPUA&list=PLQVvvaa0QuDdttJXlLtAJxJetJcqmqlQq) and many other resources._
-> _This project is dedicated to [Technothlon](https://www.facebook.com/technothlon.techniche) where the idea of making such solution was conceived. Technothlon is a logic-based examination organized by students of IIT Guwahati._
+_**Satya Mallic** for his resourceful blog:_ https://www.learnopencv.com
-
-## License
-```
-Copyright © 2019 Udayraj Deshmukh
-OMRChecker : Grade exams fast and accurately using a scanner 🖨 or your phone 🤳
-This is free software, and you are welcome to redistribute it under certain conditions;
-```
-For more details see [](https://github.com/Udayraj123/OMRChecker/blob/master/LICENSE)
-
## Related Projects
-Here's a sneak peak of the [Android OMR Helper App(WIP)](https://github.com/Udayraj123/AndroidOMRHelper):
+
+Here's a snapshot of the [Android OMR Helper App (archived)](https://github.com/Udayraj123/AndroidOMRHelper):
+
-  +
+ 
-
-### Other ways you can contribute:
-- Help OMRChecker cross 560 stars ⭐ to become #1 ([Currently #4](https://github.com/topics/omr)).
-Current stars: [](https://GitHub.com/Udayraj123/OMRChecker/stargazers/)
+## Stargazers over time
-- [Buy Me A Coffee ☕](https://www.buymeacoffee.com/Udayraj123) - To keep my brain juices flowing and help me create more such projects 💡
+[](https://starchart.cc/Udayraj123/OMRChecker)
-- If this project saved you large costs on OMR Software licenses, or saved efforts to make one. [](https://www.paypal.me/Udayraj123/500)
+---
-
-
-
+Made with ❤️ by Awesome Contributors
+
+
+  +
+
+---
+
+### License
+
+[](https://github.com/Udayraj123/OMRChecker/blob/master/LICENSE)
+
+For more details see [LICENSE](https://github.com/Udayraj123/OMRChecker/blob/master/LICENSE).
+
+### Donate
+
+
+
+
+---
+
+### License
+
+[](https://github.com/Udayraj123/OMRChecker/blob/master/LICENSE)
+
+For more details see [LICENSE](https://github.com/Udayraj123/OMRChecker/blob/master/LICENSE).
+
+### Donate
+
+ [](https://www.paypal.me/Udayraj123/500)
+
+_Find OMRChecker on_ [**_Product Hunt_**](https://www.producthunt.com/posts/omr-checker/) **|** [**_Reddit_**](https://www.reddit.com/r/computervision/comments/ccbj6f/omrchecker_grade_exams_using_python_and_opencv/) **|** [**Discord**](https://discord.gg/qFv2Vqf) **|** [**Linkedin**](https://www.linkedin.com/pulse/open-source-talks-udayraj-udayraj-deshmukh/) **|** [**goodfirstissue.dev**](https://goodfirstissue.dev/language/python) **|** [**codepeak.tech**](https://www.codepeak.tech/) **|** [**fossoverflow.dev**](https://fossoverflow.dev/projects) **|** [**Interview on Console by CodeSee**](https://console.substack.com/p/console-140) **|** [**Open Source Hub**](https://opensourcehub.io/udayraj123/omrchecker)
-
-*Find OMRChecker on* [***Product Hunt***](https://www.producthunt.com/posts/omr-checker/) **|** [***Hacker News***](https://news.ycombinator.com/item?id=20420602) **|** [***Reddit***](https://www.reddit.com/r/computervision/comments/ccbj6f/omrchecker_grade_exams_using_python_and_opencv/) **|** [***Swyya***](https://www.swyya.com/projects/omrchecker) **|** [](https://discord.gg/qFv2Vqf)
+
+
diff --git a/docs/assets/colored_output.jpg b/docs/assets/colored_output.jpg
new file mode 100644
index 00000000..3cafa473
Binary files /dev/null and b/docs/assets/colored_output.jpg differ
diff --git a/globals.py b/globals.py
deleted file mode 100644
index 5f9cd929..00000000
--- a/globals.py
+++ /dev/null
@@ -1,140 +0,0 @@
-"""
-
-Designed and Developed by-
-Udayraj Deshmukh
-https://github.com/Udayraj123
-
-"""
-
-"""
-Constants
-"""
-display_height = int(480)
-display_width = int(640)
-windowWidth = 1280
-windowHeight = 720
-
-saveMarked = 1
-saveCropped = 1
-showimglvl = 4
-saveimglvl = 0
-PRELIM_CHECKS = 0
-saveImgList = {}
-resetpos = [0, 0]
-explain = 0
-# autorotate=1
-
-BATCH_NO = 1000
-NO_MARKER_ERR = 12
-MULTI_BUBBLE_WARN = 15
-
-# name of template file
-TEMPLATE_FILE = 'template.json'
-MARKER_FILE = "omr_marker.jpg"
-
-# For preProcessing
-GAMMA_LOW = 0.7
-GAMMA_HIGH = 1.25
-
-ERODE_SUB_OFF = 1
-
-# For new ways of determining threshold
-MIN_GAP, MIN_STD = 30, 25
-MIN_JUMP = 25
-# If only not confident, take help of globalTHR
-CONFIDENT_JUMP = MIN_JUMP + 15
-JUMP_DELTA = 30
-# MIN_GAP : worst case gap of black and gray
-
-# Templ alignment parameters
-ALIGN_RANGE = range(-5, 6, 1)
-# TODO ^THIS SHOULD BE IN LAYOUT FILE AS ITS RELATED TO DIMENSIONS
-# ALIGN_RANGE = [-6,-4,-2,-1,0,1,2,4,6]
-
-# max threshold difference for template matching
-thresholdVar = 0.41
-
-# TODO: remove unnec variables here-
-thresholdCircle = 0.3
-marker_rescale_range = (35, 100)
-marker_rescale_steps = 10
-
-# Presentation variables
-uniform_height = int(1231 / 1.5)
-uniform_width = int(1000 / 1.5)
-# Original dims are about (3527, 2494)
-
-# Any input images should be resized to this--
-uniform_width_hd = int(uniform_width * 1.5)
-uniform_height_hd = int(uniform_height * 1.5)
-
-TEXT_SIZE = 0.95
-CLR_BLACK = (50, 150, 150)
-CLR_WHITE = (250, 250, 250)
-CLR_GRAY = (130, 130, 130)
-# CLR_DARK_GRAY = (190,190,190)
-CLR_DARK_GRAY = (100, 100, 100)
-
-MIN_PAGE_AREA = 80000
-
-# Filepaths
-
-
-class Paths:
- def __init__(self, output):
- self.output = output
- self.saveMarkedDir = f'{output}/CheckedOMRs/'
- self.resultDir = f'{output}/Results/'
- self.manualDir = f'{output}/Manual/'
- self.errorsDir = f'{self.manualDir}ErrorFiles/'
- self.badRollsDir = f'{self.manualDir}BadRollNosFiles/'
- self.multiMarkedDir = f'{self.manualDir}MultiMarkedFiles/'
-
-
-"""
-Variables
-"""
-filesMoved = 0
-filesNotMoved = 0
-
-# for positioning image windows
-windowX, windowY = 0, 0
-
-
-# TODO: move to template or similar json
-Answers = {
- 'J': {
- 'q1': ['B'], 'q2': ['B'], 'q3': ['B'], 'q4': ['C'], 'q5': ['0', '00'], 'q6': ['0', '00'], 'q7': ['4', '04'],
- 'q8': ['9', '09'], 'q9': ['11', '11'], 'q10': ['C'], 'q11': ['C'], 'q12': ['B'], 'q13': ['C'],
- 'q14': ['C'], 'q15': ['B'], 'q16': ['C'], 'q17': ['BONUS'], 'q18': ['A'], 'q19': ['C'], 'q20': ['B']},
- 'H': {
- 'q1': ['B'], 'q2': ['BONUS'], 'q3': ['A'], 'q4': ['B'], 'q5': ['A'], 'q6': ['B'], 'q7': ['B'],
- 'q8': ['C'], 'q9': ['4', '04'], 'q10': ['4', '04'], 'q11': ['5', '05'], 'q12': ['1', '01'], 'q13': ['28'],
- 'q14': ['C'], 'q15': ['B'], 'q16': ['C'], 'q17': ['C'], 'q18': ['C'], 'q19': ['B'], 'q20': ['C']},
- 'JK': {
- 'q1': ['B'], 'q2': ['B'], 'q3': ['B'], 'q4': ['C'], 'q5': ['0', '00'], 'q6': ['0', '00'], 'q7': ['4', '04'],
- 'q8': ['9', '09'], 'q9': ['11', '11'], 'q10': ['C'], 'q11': ['C'], 'q12': ['B'], 'q13': ['C'],
- 'q14': ['C'], 'q15': ['B'], 'q16': ['C'], 'q17': ['BONUS'], 'q18': ['A'], 'q19': ['C'], 'q20': ['B']},
- 'HK': {
- 'q1': ['B'], 'q2': ['BONUS'], 'q3': ['A'], 'q4': ['B'], 'q5': ['B'], 'q6': ['B'], 'q7': ['B'],
- 'q8': ['C'], 'q9': ['4', '04'], 'q10': ['4', '04'], 'q11': ['5', '05'], 'q12': ['1', '01'], 'q13': ['28'],
- 'q14': ['C'], 'q15': ['B'], 'q16': ['C'], 'q17': ['C'], 'q18': ['C'], 'q19': ['B'], 'q20': ['C']},
-}
-
-# TODO: Make this generalized and move it to samples
-Sections = {
- 'J': {
- 'Fibo1': {'ques': [1, 2, 3, 4], '+seq': [2, 3, 5, 8], '-seq': [0, 1, 1, 2]},
- 'Power1': {'ques': [5, 6, 7, 8, 9], '+seq': [1, 2, 4, 8, 16], '-seq': [0, 0, 0, 0, 0]},
- 'Fibo2': {'ques': [10, 11, 12, 13], '+seq': [2, 3, 5, 8], '-seq': [0, 1, 1, 2]},
- 'allNone1': {'ques': [14, 15, 16], 'marks': 12},
- 'Boom1': {'ques': [17, 18, 19, 20], '+seq': [3, 3, 3, 3], '-seq': [1, 1, 1, 1]},
- },
- 'H': {
- 'Boom1': {'ques': [1, 2, 3, 4], '+seq': [3, 3, 3, 3], '-seq': [1, 1, 1, 1]},
- 'Fibo1': {'ques': [5, 6, 7, 8], '+seq': [2, 3, 5, 8], '-seq': [0, 1, 1, 2]},
- 'Power1': {'ques': [9, 10, 11, 12, 13], '+seq': [1, 2, 4, 8, 16], '-seq': [0, 0, 0, 0, 0]},
- 'allNone1': {'ques': [14, 15, 16], 'marks': 12},
- 'Boom2': {'ques': [17, 18, 19, 20], '+seq': [3, 3, 3, 3], '-seq': [1, 1, 1, 1]},
- },
-}
diff --git a/main.py b/main.py
index eac70a50..dfecbca3 100644
--- a/main.py
+++ b/main.py
@@ -1,546 +1,99 @@
"""
-Designed and Developed by-
-Udayraj Deshmukh
-https://github.com/Udayraj123
+ OMRChecker
+
+ Author: Udayraj Deshmukh
+ Github: https://github.com/Udayraj123
"""
-import re
-import os
-import cv2
import argparse
-import numpy as np
-import pandas as pd
-import matplotlib.pyplot as plt
-
-# from utils import * #Now imported via template
-from globals import *
-from template import *
-from glob import glob
-from csv import QUOTE_NONNUMERIC
-from time import localtime, strftime, time
-
-
-# TODO(beginner task) :-
-# from colorama import init
-# init()
-# from colorama import Fore, Back, Style
-
-def process_dir(root_dir, subdir, template):
- curr_dir = os.path.join(root_dir, subdir)
-
- # Look for template in current dir
- template_file = os.path.join(curr_dir, TEMPLATE_FILE)
- if os.path.exists(template_file):
- template = Template(template_file)
-
- # look for images in current dir to process
- paths = Paths(os.path.join(args['output_dir'], subdir))
- exts = ('*.png', '*.jpg')
- omr_files = sorted(
- [f for ext in exts for f in glob(os.path.join(curr_dir, ext))])
-
- # Exclude marker image if exists
- if(template and template.marker_path):
- omr_files = [f for f in omr_files if f != template.marker_path]
-
- subfolders = sorted([file for file in os.listdir(
- curr_dir) if os.path.isdir(os.path.join(curr_dir, file))])
- if omr_files:
- args_local = args.copy()
- if("OverrideFlags" in template.options):
- args_local.update(template.options["OverrideFlags"])
- print('\n------------------------------------------------------------------')
- print(f'Processing directory "{curr_dir}" with settings- ')
- print("\tTotal images : %d" % (len(omr_files)))
- print("\tCropping Enabled : " + str(not args_local["noCropping"]))
- print("\tAuto Alignment : " + str(args_local["autoAlign"]))
- print("\tUsing Template : " + str(template.path) if(template) else "N/A")
- print("\tUsing Marker : " + str(template.marker_path)
- if(template.marker is not None) else "N/A")
- print('')
-
- if not template:
- print(f'Error: No template file when processing {curr_dir}.')
- print(
- f' Place {TEMPLATE_FILE} in the directory or specify a template using -t.')
- return
-
- setup_dirs(paths)
- output_set = setup_output(paths, template)
- process_files(omr_files, template, args_local, output_set)
- elif(len(subfolders) == 0):
- # the directory should have images or be non-leaf
- print(f'Note: No valid images or subfolders found in {curr_dir}')
-
- # recursively process subfolders
- for folder in subfolders:
- process_dir(root_dir, os.path.join(subdir, folder), template)
-
-
-def checkAndMove(error_code, filepath, filepath2):
- # print("Dummy Move: "+filepath, " --> ",filepath2)
- global filesNotMoved
- filesNotMoved += 1
- return True
-
- global filesMoved
- if(not os.path.exists(filepath)):
- print('File already moved')
- return False
- if(os.path.exists(filepath2)):
- print('ERROR : Duplicate file at ' + filepath2)
- return False
-
- print("Moved: " + filepath, " --> ", filepath2)
- os.rename(filepath, filepath2)
- filesMoved += 1
- return True
-
-
-def processOMR(template, omrResp):
- # Note: This is a reference function. It is not part of the OMR checker
- # So its implementation is completely subjective to user's requirements.
- csvResp = {}
-
- # symbol for absent response
- UNMARKED_SYMBOL = ''
-
- # print("omrResp",omrResp)
-
- # Multi-column/multi-row questions which need to be concatenated
- for qNo, respKeys in template.concats.items():
- csvResp[qNo] = ''.join([omrResp.get(k, UNMARKED_SYMBOL)
- for k in respKeys])
-
- # Single-column/single-row questions
- for qNo in template.singles:
- csvResp[qNo] = omrResp.get(qNo, UNMARKED_SYMBOL)
-
- # Note: Concatenations and Singles together should be mutually exclusive
- # and should cover all questions in the template(exhaustive)
- # TODO: ^add a warning if omrResp has unused keys remaining
- return csvResp
-
-
-def report(
- Status,
- streak,
- scheme,
- qNo,
- marked,
- ans,
- prevmarks,
- currmarks,
- marks):
- print(
- '%s \t %s \t\t %s \t %s \t %s \t %s \t %s ' % (qNo,
- Status,
- str(streak),
- '[' + scheme + '] ',
- (str(prevmarks) + ' + ' + str(currmarks) + ' =' + str(marks)),
- str(marked),
- str(ans)))
-
-# check sectionwise only.
-
-
-def evaluate(resp, squad="H", explain=False):
- # TODO: @contributors - Need help generalizing this function
- global Answers, Sections
- marks = 0
- answers = Answers[squad]
- if(explain):
- print('Question\tStatus \t Streak\tSection \tMarks_Update\tMarked:\tAnswer:')
- for scheme, section in Sections[squad].items():
- sectionques = section['ques']
- prevcorrect = None
- allflag = 1
- streak = 0
- for q in sectionques:
- qNo = 'q' + str(q)
- ans = answers[qNo]
- marked = resp.get(qNo, 'X')
- firstQ = sectionques[0]
- lastQ = sectionques[len(sectionques) - 1]
- unmarked = marked == 'X' or marked == ''
- bonus = 'BONUS' in ans
- correct = bonus or (marked in ans)
- inrange = 0
-
- if(unmarked or int(q) == firstQ):
- streak = 0
- elif(prevcorrect == correct):
- streak += 1
- else:
- streak = 0
-
- if('allNone' in scheme):
- # loop on all sectionques
- allflag = allflag and correct
- if(q == lastQ):
- # at the end check allflag
- prevcorrect = correct
- currmarks = section['marks'] if allflag else 0
- else:
- currmarks = 0
-
- elif('Proxy' in scheme):
- a = int(ans[0])
- # proximity check
- inrange = 1 if unmarked else (
- float(abs(int(marked) - a)) / float(a) <= 0.25)
- currmarks = section['+marks'] if correct else (
- 0 if inrange else -section['-marks'])
-
- elif('Fibo' in scheme or 'Power' in scheme or 'Boom' in scheme):
- currmarks = section['+seq'][streak] if correct else (
- 0 if unmarked else -section['-seq'][streak])
- elif('TechnoFin' in scheme):
- currmarks = 0
- else:
- print('Invalid Sections')
- prevmarks = marks
- marks += currmarks
-
- if(explain):
- if bonus:
- report('BonusQ', streak, scheme, qNo, marked,
- ans, prevmarks, currmarks, marks)
- elif correct:
- report('Correct', streak, scheme, qNo, marked,
- ans, prevmarks, currmarks, marks)
- elif unmarked:
- report('Unmarked', streak, scheme, qNo, marked,
- ans, prevmarks, currmarks, marks)
- elif inrange:
- report('InProximity', streak, scheme, qNo,
- marked, ans, prevmarks, currmarks, marks)
- else:
- report('Incorrect', streak, scheme, qNo,
- marked, ans, prevmarks, currmarks, marks)
-
- prevcorrect = correct
-
- return marks
-
-
-def setup_output(paths, template):
- ns = argparse.Namespace()
- print("\nChecking Files...")
-
- # Include current output paths
- ns.paths = paths
-
- # custom sort: To use integer order in question names instead of
- # alphabetical - avoids q1, q10, q2 and orders them q1, q2, ..., q10
- ns.respCols = sorted(list(template.concats.keys()) + template.singles,
- key=lambda x: int(x[1:]) if ord(x[1]) in range(48, 58) else 0)
- ns.emptyResp = [''] * len(ns.respCols)
- ns.sheetCols = ['file_id', 'input_path',
- 'output_path', 'score'] + ns.respCols
- ns.OUTPUT_SET = []
- ns.filesObj = {}
- ns.filesMap = {
- "Results": paths.resultDir + 'Results_' + timeNowHrs + '.csv',
- "MultiMarked": paths.manualDir + 'MultiMarkedFiles_.csv',
- "Errors": paths.manualDir + 'ErrorFiles_.csv',
- "BadRollNos": paths.manualDir + 'BadRollNoFiles_.csv'
- }
-
- for fileKey, fileName in ns.filesMap.items():
- if(not os.path.exists(fileName)):
- print("Note: Created new file: %s" % (fileName))
- # still append mode req [THINK!]
- ns.filesObj[fileKey] = open(fileName, 'a')
- # Create Header Columns
- pd.DataFrame([ns.sheetCols], dtype=str).to_csv(
- ns.filesObj[fileKey], quoting=QUOTE_NONNUMERIC, header=False, index=False)
- else:
- print('Present : appending to %s' % (fileName))
- ns.filesObj[fileKey] = open(fileName, 'a')
-
- return ns
-
-
-''' TODO: Refactor into new process flow.
- Currently I have no idea what this does so I left it out'''
-
-
-def preliminary_check():
- filesCounter = 0
- # PRELIM_CHECKS for thresholding
- if(PRELIM_CHECKS):
- # TODO: add more using unit testing
- ALL_WHITE = 255 * \
- np.ones((TEMPLATE.dims[1], TEMPLATE.dims[0]), dtype='uint8')
- OMRresponseDict, final_marked, MultiMarked, multiroll = readResponse(
- ALL_WHITE, name="ALL_WHITE", savedir=None, autoAlign=True)
- print("ALL_WHITE", OMRresponseDict)
- if(OMRresponseDict != {}):
- print("Preliminary Checks Failed.")
- exit(12)
- ALL_BLACK = np.zeros(
- (TEMPLATE.dims[1], TEMPLATE.dims[0]), dtype='uint8')
- OMRresponseDict, final_marked, MultiMarked, multiroll = readResponse(
- ALL_BLACK, name="ALL_BLACK", savedir=None, autoAlign=True)
- print("ALL_BLACK", OMRresponseDict)
- show("Confirm : All bubbles are black", final_marked, 1, 1)
-
-
-def process_files(omr_files, template, args, out):
- start_time = int(time())
- filesCounter = 0
- filesNotMoved = 0
-
- for filepath in omr_files:
- filesCounter += 1
- # For windows filesystem support: all '\' will be replaced by '/'
- filepath = filepath.replace(os.sep, '/')
-
- # Prefixing a 'r' to use raw string (escape character '\' is taken
- # literally)
- finder = re.search(r'.*/(.*)/(.*)', filepath, re.IGNORECASE)
- if(finder):
- inputFolderName, filename = finder.groups()
- else:
- print("Error: Filepath not matching to Regex: " + filepath)
- continue
- # set global var for reading
-
- inOMR = cv2.imread(filepath, cv2.IMREAD_GRAYSCALE)
- print(
- '\n[%d] Processing image: \t' %
- (filesCounter),
- filepath,
- "\tResolution: ",
- inOMR.shape)
-
- OMRCrop = getROI(inOMR, filename, noCropping=args["noCropping"])
-
- if(OMRCrop is None):
- # Error OMR - could not crop
- newfilepath = out.paths.errorsDir + filename
- out.OUTPUT_SET.append([filename] + out.emptyResp)
- if(checkAndMove(NO_MARKER_ERR, filepath, newfilepath)):
- err_line = [filename, filepath,
- newfilepath, "NA"] + out.emptyResp
- pd.DataFrame(
- err_line,
- dtype=str).T.to_csv(
- out.filesObj["Errors"],
- quoting=QUOTE_NONNUMERIC,
- header=False,
- index=False)
- continue
-
- if template.marker is not None:
- OMRCrop = handle_markers(OMRCrop, template.marker, filename)
-
- if(args["setLayout"]):
- templateLayout = drawTemplateLayout(
- OMRCrop, template, shifted=False, border=2)
- show("Template Layout", templateLayout, 1, 1)
- continue
-
- # uniquify
- file_id = inputFolderName + '_' + filename
- savedir = out.paths.saveMarkedDir
- OMRresponseDict, final_marked, MultiMarked, multiroll = \
- readResponse(template, OMRCrop, name=file_id,
- savedir=savedir, autoAlign=args["autoAlign"])
-
- # concatenate roll nos, set unmarked responses, etc
- resp = processOMR(template, OMRresponseDict)
- print("\nRead Response: \t", resp)
-
- # This evaluates and returns the score attribute
- score = evaluate(resp, explain=explain)
- respArray = []
- for k in out.respCols:
- respArray.append(resp[k])
-
- out.OUTPUT_SET.append([filename] + respArray)
-
- # TODO: Add roll number validation here
- if(MultiMarked == 0):
- filesNotMoved += 1
- newfilepath = savedir + file_id
- # Enter into Results sheet-
- results_line = [filename, filepath, newfilepath, score] + respArray
- # Write/Append to results_line file(opened in append mode)
- pd.DataFrame(
- results_line,
- dtype=str).T.to_csv(
- out.filesObj["Results"],
- quoting=QUOTE_NONNUMERIC,
- header=False,
- index=False)
- print("[%d] Graded with score: %.2f" %
- (filesCounter, score), '\t file_id: ', file_id)
- # print(filesCounter,file_id,resp['Roll'],'score : ',score)
- else:
- # MultiMarked file
- print('[%d] MultiMarked, moving File: %s' %
- (filesCounter, file_id))
- newfilepath = out.paths.multiMarkedDir + filename
- if(checkAndMove(MULTI_BUBBLE_WARN, filepath, newfilepath)):
- mm_line = [filename, filepath, newfilepath, "NA"] + respArray
- pd.DataFrame(
- mm_line,
- dtype=str).T.to_csv(
- out.filesObj["MultiMarked"],
- quoting=QUOTE_NONNUMERIC,
- header=False,
- index=False)
- # else:
- # TODO: Add appropriate record handling here
- # pass
-
- # flush after every 20 files for a live view
- if(filesCounter % 20 == 0 or filesCounter == len(omr_files)):
- for fileKey in out.filesMap.keys():
- out.filesObj[fileKey].flush()
-
- timeChecking = round(time() - start_time, 2) if filesCounter else 1
- print('')
- print('Total files moved : %d ' % (filesMoved))
- print('Total files not moved : %d ' % (filesNotMoved))
- print('------------------------------')
- print(
- 'Total files processed : %d (%s)' %
- (filesCounter,
- 'Sum Tallied!' if filesCounter == (
- filesMoved +
- filesNotMoved) else 'Not Tallying!'))
-
- if(showimglvl <= 0):
- print(
- '\nFinished Checking %d files in %.1f seconds i.e. ~%.1f minutes.' %
- (filesCounter, timeChecking, timeChecking / 60))
- print('OMR Processing Rate :\t ~ %.2f seconds/OMR' %
- (timeChecking / filesCounter))
- print('OMR Processing Speed :\t ~ %.2f OMRs/minute' %
- ((filesCounter * 60) / timeChecking))
- else:
- print("\nTotal script time :", timeChecking, "seconds")
-
- if(showimglvl <= 1):
- # TODO: colorama this
- print(
- "\nTip: To see some awesome visuals, open globals.py and increase 'showimglvl'")
-
- evaluate_correctness(template, out)
-
- # Use this data to train as +ve feedback
- if(showimglvl >= 0 and filesCounter > 10):
- # TODO: Find good parameters to plot and depict image set quality
- for x in [thresholdCircles]: # ,badThresholds,veryBadPoints, , mbs]:
- if(x != []):
- x = pd.DataFrame(x)
- print(x.describe())
- plt.plot(range(len(x)), x)
- plt.title("Mystery Plot")
- plt.show()
- else:
- print(x)
-
-
-# Evaluate accuracy based on OMRDataset file generated through moderation
-# portal on the same set of images
-def evaluate_correctness(template, out):
- # TODO: TEST_FILE WOULD BE RELATIVE TO INPUT SUBDIRECTORY NOW-
- TEST_FILE = 'inputs/OMRDataset.csv'
- if(os.path.exists(TEST_FILE)):
- print("\nStarting evaluation for: " + TEST_FILE)
-
- TEST_COLS = ['file_id'] + out.respCols
- y_df = pd.read_csv(
- TEST_FILE, dtype=str)[TEST_COLS].replace(
- np.nan, '', regex=True).set_index('file_id')
-
- if(np.any(y_df.index.duplicated)):
- y_df_filtered = y_df.loc[~y_df.index.duplicated(keep='first')]
- print(
- "WARNING: Found duplicate File-ids in file %s. Removed %d rows from testing data. Rows remaining: %d" %
- (TEST_FILE, y_df.shape[0] - y_df_filtered.shape[0], y_df_filtered.shape[0]))
- y_df = y_df_filtered
-
- x_df = pd.DataFrame(
- out.OUTPUT_SET,
- dtype=str,
- columns=TEST_COLS).set_index('file_id')
- # print("x_df",x_df.head())
- # print("\ny_df",y_df.head())
- intersection = y_df.index.intersection(x_df.index)
-
- # Checking if the merge is okay
- if(intersection.size == x_df.index.size):
- y_df = y_df.loc[intersection]
- x_df['TestResult'] = (x_df == y_df).all(axis=1).astype(int)
- print(x_df.head())
- print("\n\t Accuracy on the %s Dataset: %.6f" %
- (TEST_FILE, (x_df['TestResult'].sum() / x_df.shape[0])))
- else:
- print(
- "\nERROR: Insufficient Testing Data: Have you appended MultiMarked data yet?")
- print("Missing File-ids: ",
- list(x_df.index.difference(intersection)))
-
-
-timeNowHrs = strftime("%I%p", localtime())
-
-# construct the argument parse and parse the arguments
-argparser = argparse.ArgumentParser()
-# https://docs.python.org/3/howto/argparse.html
-# store_true: if the option is specified, assign the value True to
-# args.verbose. Not specifying it implies False.
-argparser.add_argument(
- "-c",
- "--noCropping",
- required=False,
- dest='noCropping',
- action='store_true',
- help="Disables page contour detection - used when page boundary is not visible e.g. document scanner.")
-argparser.add_argument(
- "-a",
- "--autoAlign",
- required=False,
- dest='autoAlign',
- action='store_true',
- help="(experimental) Enables automatic template alignment - use if the scans show slight misalignments.")
-argparser.add_argument(
- "-l",
- "--setLayout",
- required=False,
- dest='setLayout',
- action='store_true',
- help="Set up OMR template layout - modify your json file and run again until the template is set.")
-argparser.add_argument("-i", "--inputDir", required=False, action='append',
- dest='input_dir', help="Specify an input directory.")
-argparser.add_argument("-o", "--outputDir", default='outputs', required=False,
- dest='output_dir', help="Specify an output directory.")
-argparser.add_argument(
- "-t",
- "--template",
- required=False,
- dest='template',
- help="Specify a default template if no template file in input directories.")
-
-
-args, unknown = argparser.parse_known_args()
-args = vars(args)
-if(len(unknown) > 0):
- print("\nError: Unknown arguments:", unknown)
- argparser.print_help()
- exit(11)
-
-if args['template']:
- args['template'] = Template(args['template'])
-
-if args['input_dir'] is None:
- args['input_dir'] = ['inputs']
-
-for root in args['input_dir']:
- process_dir(root, '', args['template'])
+import sys
+from pathlib import Path
+
+from src.entry import entry_point
+from src.logger import logger
+
+
+def parse_args():
+ # construct the argument parse and parse the arguments
+ argparser = argparse.ArgumentParser()
+
+ argparser.add_argument(
+ "-i",
+ "--inputDir",
+ default=["inputs"],
+ # https://docs.python.org/3/library/argparse.html#nargs
+ nargs="*",
+ required=False,
+ type=str,
+ dest="input_paths",
+ help="Specify an input directory.",
+ )
+
+ argparser.add_argument(
+ "-d",
+ "--debug",
+ required=False,
+ dest="debug",
+ action="store_false",
+ help="Enables debugging mode for showing detailed errors",
+ )
+
+ argparser.add_argument(
+ "-o",
+ "--outputDir",
+ default="outputs",
+ required=False,
+ dest="output_dir",
+ help="Specify an output directory.",
+ )
+
+ argparser.add_argument(
+ "-a",
+ "--autoAlign",
+ required=False,
+ dest="autoAlign",
+ action="store_true",
+ help="(experimental) Enables automatic template alignment - \
+ use if the scans show slight misalignments.",
+ )
+
+ argparser.add_argument(
+ "-l",
+ "--setLayout",
+ required=False,
+ dest="setLayout",
+ action="store_true",
+ help="Set up OMR template layout - modify your json file and \
+ run again until the template is set.",
+ )
+
+ (
+ args,
+ unknown,
+ ) = argparser.parse_known_args()
+
+ args = vars(args)
+
+ if len(unknown) > 0:
+ logger.warning(f"\nError: Unknown arguments: {unknown}", unknown)
+ argparser.print_help()
+ exit(11)
+ return args
+
+
+def entry_point_for_args(args):
+ if args["debug"] is True:
+ # Disable tracebacks
+ sys.tracebacklimit = 0
+ for root in args["input_paths"]:
+ entry_point(
+ Path(root),
+ args,
+ )
+
+
+if __name__ == "__main__":
+ args = parse_args()
+ entry_point_for_args(args)
diff --git a/pyproject.toml b/pyproject.toml
new file mode 100644
index 00000000..060309be
--- /dev/null
+++ b/pyproject.toml
@@ -0,0 +1,18 @@
+[tool.black]
+exclude = '''
+(
+ /(
+ \.eggs # exclude a few common directories in the
+ | \.git # root of the project
+ | \.venv
+ | _build
+ | build
+ | dist
+ )/
+ | foo.py # also separately exclude a file named foo.py in
+ # the root of the project
+)
+'''
+include = '\.pyi?$'
+line-length = 88
+target-version = ['py37']
diff --git a/pytest.ini b/pytest.ini
new file mode 100644
index 00000000..84008a21
--- /dev/null
+++ b/pytest.ini
@@ -0,0 +1,6 @@
+# pytest.ini
+[pytest]
+minversion = 7.0
+addopts = -qq --capture=no
+testpaths =
+ src/tests
diff --git a/requirements.dev.txt b/requirements.dev.txt
new file mode 100644
index 00000000..722ab0dd
--- /dev/null
+++ b/requirements.dev.txt
@@ -0,0 +1,7 @@
+-r requirements.txt
+flake8>=6.0.0
+freezegun>=1.2.2
+pre-commit>=3.3.3
+pytest-mock>=3.11.1
+pytest>=7.4.0
+syrupy>=4.0.4
diff --git a/requirements.txt b/requirements.txt
index 505865c9..761d2e51 100644
--- a/requirements.txt
+++ b/requirements.txt
@@ -1,4 +1,8 @@
-imutils>=0.5.2
-matplotlib>=3.0.2
-numpy>=1.16.0
-pandas>=0.24.0
+deepmerge>=1.1.0
+dotmap>=1.3.30
+jsonschema>=4.17.3
+matplotlib>=3.7.1
+numpy>=1.25.0
+pandas>=2.0.2
+rich>=13.4.2
+screeninfo>=0.8.1
diff --git a/samples/answer-key/using-csv/adrian_omr.png b/samples/answer-key/using-csv/adrian_omr.png
new file mode 100644
index 00000000..d8db0994
Binary files /dev/null and b/samples/answer-key/using-csv/adrian_omr.png differ
diff --git a/samples/answer-key/using-csv/answer_key.csv b/samples/answer-key/using-csv/answer_key.csv
new file mode 100644
index 00000000..566201d1
--- /dev/null
+++ b/samples/answer-key/using-csv/answer_key.csv
@@ -0,0 +1,5 @@
+q1,C
+q2,E
+q3,A
+q4,B
+q5,B
\ No newline at end of file
diff --git a/samples/answer-key/using-csv/evaluation.json b/samples/answer-key/using-csv/evaluation.json
new file mode 100644
index 00000000..14a3db25
--- /dev/null
+++ b/samples/answer-key/using-csv/evaluation.json
@@ -0,0 +1,14 @@
+{
+ "source_type": "csv",
+ "options": {
+ "answer_key_csv_path": "answer_key.csv",
+ "should_explain_scoring": true
+ },
+ "marking_schemes": {
+ "DEFAULT": {
+ "correct": "1",
+ "incorrect": "0",
+ "unmarked": "0"
+ }
+ }
+}
diff --git a/samples/answer-key/using-csv/template.json b/samples/answer-key/using-csv/template.json
new file mode 100644
index 00000000..41ec9ffa
--- /dev/null
+++ b/samples/answer-key/using-csv/template.json
@@ -0,0 +1,35 @@
+{
+ "pageDimensions": [
+ 300,
+ 400
+ ],
+ "bubbleDimensions": [
+ 25,
+ 25
+ ],
+ "preProcessors": [
+ {
+ "name": "CropPage",
+ "options": {
+ "morphKernel": [
+ 10,
+ 10
+ ]
+ }
+ }
+ ],
+ "fieldBlocks": {
+ "MCQ_Block_1": {
+ "fieldType": "QTYPE_MCQ5",
+ "origin": [
+ 65,
+ 60
+ ],
+ "fieldLabels": [
+ "q1..5"
+ ],
+ "labelsGap": 52,
+ "bubblesGap": 41
+ }
+ }
+}
diff --git a/samples/answer-key/weighted-answers/evaluation.json b/samples/answer-key/weighted-answers/evaluation.json
new file mode 100644
index 00000000..c0daefcf
--- /dev/null
+++ b/samples/answer-key/weighted-answers/evaluation.json
@@ -0,0 +1,35 @@
+{
+ "source_type": "custom",
+ "options": {
+ "questions_in_order": [
+ "q1..5"
+ ],
+ "answers_in_order": [
+ "C",
+ "E",
+ [
+ "A",
+ "C"
+ ],
+ [
+ [
+ "B",
+ 2
+ ],
+ [
+ "C",
+ "3/2"
+ ]
+ ],
+ "C"
+ ],
+ "should_explain_scoring": true
+ },
+ "marking_schemes": {
+ "DEFAULT": {
+ "correct": "3",
+ "incorrect": "-1",
+ "unmarked": "0"
+ }
+ }
+}
diff --git a/samples/answer-key/weighted-answers/images/adrian_omr.png b/samples/answer-key/weighted-answers/images/adrian_omr.png
new file mode 100644
index 00000000..69a53823
Binary files /dev/null and b/samples/answer-key/weighted-answers/images/adrian_omr.png differ
diff --git a/samples/answer-key/weighted-answers/images/adrian_omr_2.png b/samples/answer-key/weighted-answers/images/adrian_omr_2.png
new file mode 100644
index 00000000..d8db0994
Binary files /dev/null and b/samples/answer-key/weighted-answers/images/adrian_omr_2.png differ
diff --git a/samples/answer-key/weighted-answers/template.json b/samples/answer-key/weighted-answers/template.json
new file mode 100644
index 00000000..41ec9ffa
--- /dev/null
+++ b/samples/answer-key/weighted-answers/template.json
@@ -0,0 +1,35 @@
+{
+ "pageDimensions": [
+ 300,
+ 400
+ ],
+ "bubbleDimensions": [
+ 25,
+ 25
+ ],
+ "preProcessors": [
+ {
+ "name": "CropPage",
+ "options": {
+ "morphKernel": [
+ 10,
+ 10
+ ]
+ }
+ }
+ ],
+ "fieldBlocks": {
+ "MCQ_Block_1": {
+ "fieldType": "QTYPE_MCQ5",
+ "origin": [
+ 65,
+ 60
+ ],
+ "fieldLabels": [
+ "q1..5"
+ ],
+ "labelsGap": 52,
+ "bubblesGap": 41
+ }
+ }
+}
diff --git a/samples/community/Antibodyy/simple_omr_sheet.jpg b/samples/community/Antibodyy/simple_omr_sheet.jpg
new file mode 100644
index 00000000..661d5f4f
Binary files /dev/null and b/samples/community/Antibodyy/simple_omr_sheet.jpg differ
diff --git a/samples/community/Antibodyy/template.json b/samples/community/Antibodyy/template.json
new file mode 100644
index 00000000..7e962bbf
--- /dev/null
+++ b/samples/community/Antibodyy/template.json
@@ -0,0 +1,35 @@
+{
+ "pageDimensions": [
+ 299,
+ 398
+ ],
+ "bubbleDimensions": [
+ 42,
+ 42
+ ],
+ "fieldBlocks": {
+ "MCQBlock1": {
+ "fieldType": "QTYPE_MCQ5",
+ "origin": [
+ 65,
+ 79
+ ],
+ "bubblesGap": 43,
+ "labelsGap": 50,
+ "fieldLabels": [
+ "q1..6"
+ ]
+ }
+ },
+ "preProcessors": [
+ {
+ "name": "CropPage",
+ "options": {
+ "morphKernel": [
+ 10,
+ 10
+ ]
+ }
+ }
+ ]
+}
diff --git a/samples/community/Sandeep-1507/omr-1.png b/samples/community/Sandeep-1507/omr-1.png
new file mode 100644
index 00000000..8c40655e
Binary files /dev/null and b/samples/community/Sandeep-1507/omr-1.png differ
diff --git a/samples/community/Sandeep-1507/omr-2.png b/samples/community/Sandeep-1507/omr-2.png
new file mode 100644
index 00000000..aabef0a5
Binary files /dev/null and b/samples/community/Sandeep-1507/omr-2.png differ
diff --git a/samples/community/Sandeep-1507/omr-3.png b/samples/community/Sandeep-1507/omr-3.png
new file mode 100644
index 00000000..8f1cb5c8
Binary files /dev/null and b/samples/community/Sandeep-1507/omr-3.png differ
diff --git a/samples/community/Sandeep-1507/template.json b/samples/community/Sandeep-1507/template.json
new file mode 100644
index 00000000..b35404e2
--- /dev/null
+++ b/samples/community/Sandeep-1507/template.json
@@ -0,0 +1,234 @@
+{
+ "pageDimensions": [
+ 1189,
+ 1682
+ ],
+ "bubbleDimensions": [
+ 15,
+ 15
+ ],
+ "preProcessors": [
+ {
+ "name": "GaussianBlur",
+ "options": {
+ "kSize": [
+ 3,
+ 3
+ ],
+ "sigmaX": 0
+ }
+ }
+ ],
+ "customLabels": {
+ "Booklet_No": [
+ "b1..7"
+ ]
+ },
+ "fieldBlocks": {
+ "Booklet_No": {
+ "fieldType": "QTYPE_INT",
+ "origin": [
+ 112,
+ 530
+ ],
+ "fieldLabels": [
+ "b1..7"
+ ],
+ "emptyValue": "no",
+ "bubblesGap": 28,
+ "labelsGap": 26.5
+ },
+ "MCQBlock1a1": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q1..10"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 476,
+ 100
+ ]
+ },
+ "MCQBlock1a2": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q11..20"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 476,
+ 370
+ ]
+ },
+ "MCQBlock1a3": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q21..35"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 476,
+ 638
+ ]
+ },
+ "MCQBlock2a1": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q51..60"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 645,
+ 100
+ ]
+ },
+ "MCQBlock2a2": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q61..70"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 645,
+ 370
+ ]
+ },
+ "MCQBlock2a3": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q71..85"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 645,
+ 638
+ ]
+ },
+ "MCQBlock3a1": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q101..110"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 815,
+ 100
+ ]
+ },
+ "MCQBlock3a2": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q111..120"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 815,
+ 370
+ ]
+ },
+ "MCQBlock3a3": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q121..135"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 815,
+ 638
+ ]
+ },
+ "MCQBlock4a1": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q151..160"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 983,
+ 100
+ ]
+ },
+ "MCQBlock4a2": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q161..170"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 983,
+ 370
+ ]
+ },
+ "MCQBlock4a3": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q171..185"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 983,
+ 638
+ ]
+ },
+ "MCQBlock1a": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q36..50"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 480,
+ 1061
+ ]
+ },
+ "MCQBlock2a": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q86..100"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 648,
+ 1061
+ ]
+ },
+ "MCQBlock3a": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q136..150"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 815,
+ 1061
+ ]
+ },
+ "MCQBlock4a": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q186..200"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.6,
+ "origin": [
+ 986,
+ 1061
+ ]
+ }
+ }
+}
diff --git a/samples/community/Shamanth/omr_sheet_01.png b/samples/community/Shamanth/omr_sheet_01.png
new file mode 100644
index 00000000..ead17dd1
Binary files /dev/null and b/samples/community/Shamanth/omr_sheet_01.png differ
diff --git a/samples/community/Shamanth/template.json b/samples/community/Shamanth/template.json
new file mode 100644
index 00000000..fb18d975
--- /dev/null
+++ b/samples/community/Shamanth/template.json
@@ -0,0 +1,25 @@
+{
+ "pageDimensions": [
+ 300,
+ 400
+ ],
+ "bubbleDimensions": [
+ 20,
+ 20
+ ],
+ "fieldBlocks": {
+ "MCQBlock1": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 78,
+ 41
+ ],
+ "fieldLabels": [
+ "q21..28"
+ ],
+ "bubblesGap": 56,
+ "labelsGap": 46
+ }
+ },
+ "preProcessors": []
+}
diff --git a/samples/community/UPSC-mock/answer_key.jpg b/samples/community/UPSC-mock/answer_key.jpg
new file mode 100644
index 00000000..7a229c1b
Binary files /dev/null and b/samples/community/UPSC-mock/answer_key.jpg differ
diff --git a/samples/community/UPSC-mock/config.json b/samples/community/UPSC-mock/config.json
new file mode 100644
index 00000000..0edf9d22
--- /dev/null
+++ b/samples/community/UPSC-mock/config.json
@@ -0,0 +1,11 @@
+{
+ "dimensions": {
+ "display_height": 1800,
+ "display_width": 2400,
+ "processing_height": 2400,
+ "processing_width": 1800
+ },
+ "outputs": {
+ "show_image_level": 0
+ }
+}
diff --git a/samples/community/UPSC-mock/evaluation.json b/samples/community/UPSC-mock/evaluation.json
new file mode 100644
index 00000000..42fbec8b
--- /dev/null
+++ b/samples/community/UPSC-mock/evaluation.json
@@ -0,0 +1,18 @@
+{
+ "source_type": "csv",
+ "options": {
+ "answer_key_csv_path": "answer_key.csv",
+ "answer_key_image_path": "answer_key.jpg",
+ "questions_in_order": [
+ "q1..100"
+ ],
+ "should_explain_scoring": true
+ },
+ "marking_schemes": {
+ "DEFAULT": {
+ "correct": "2",
+ "incorrect": "-2/3",
+ "unmarked": "0"
+ }
+ }
+}
diff --git a/samples/community/UPSC-mock/scan-angles/angle-1.jpg b/samples/community/UPSC-mock/scan-angles/angle-1.jpg
new file mode 100644
index 00000000..9c75d7be
Binary files /dev/null and b/samples/community/UPSC-mock/scan-angles/angle-1.jpg differ
diff --git a/samples/community/UPSC-mock/scan-angles/angle-2.jpg b/samples/community/UPSC-mock/scan-angles/angle-2.jpg
new file mode 100644
index 00000000..edab6060
Binary files /dev/null and b/samples/community/UPSC-mock/scan-angles/angle-2.jpg differ
diff --git a/samples/community/UPSC-mock/scan-angles/angle-3.jpg b/samples/community/UPSC-mock/scan-angles/angle-3.jpg
new file mode 100644
index 00000000..f1da4af9
Binary files /dev/null and b/samples/community/UPSC-mock/scan-angles/angle-3.jpg differ
diff --git a/samples/community/UPSC-mock/template.json b/samples/community/UPSC-mock/template.json
new file mode 100644
index 00000000..a1ef5c6b
--- /dev/null
+++ b/samples/community/UPSC-mock/template.json
@@ -0,0 +1,195 @@
+{
+ "pageDimensions": [
+ 1800,
+ 2400
+ ],
+ "bubbleDimensions": [
+ 30,
+ 25
+ ],
+ "customLabels": {
+ "Subject Code": [
+ "subjectCode1",
+ "subjectCode2"
+ ],
+ "Roll": [
+ "roll1..10"
+ ]
+ },
+ "fieldBlocks": {
+ "bookletNo": {
+ "origin": [
+ 595,
+ 545
+ ],
+ "bubblesGap": 68,
+ "labelsGap": 0,
+ "fieldLabels": [
+ "bookletNo"
+ ],
+ "bubbleValues": [
+ "A",
+ "B",
+ "C",
+ "D"
+ ],
+ "direction": "vertical"
+ },
+ "subjectCode": {
+ "origin": [
+ 912,
+ 512
+ ],
+ "bubblesGap": 33,
+ "labelsGap": 42.5,
+ "fieldLabels": [
+ "subjectCode1",
+ "subjectCode2"
+ ],
+ "fieldType": "QTYPE_INT"
+ },
+ "roll": {
+ "origin": [
+ 1200,
+ 510
+ ],
+ "bubblesGap": 33,
+ "labelsGap": 42.8,
+ "fieldLabels": [
+ "roll1..10"
+ ],
+ "fieldType": "QTYPE_INT"
+ },
+ "q01block": {
+ "origin": [
+ 500,
+ 927
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q1..10"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q11block": {
+ "origin": [
+ 500,
+ 1258
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q11..20"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q21block": {
+ "origin": [
+ 500,
+ 1589
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q21..30"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q31block": {
+ "origin": [
+ 495,
+ 1925
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q31..40"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q41block": {
+ "origin": [
+ 811,
+ 927
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q41..50"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q51block": {
+ "origin": [
+ 811,
+ 1258
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q51..60"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q61block": {
+ "origin": [
+ 811,
+ 1589
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q61..70"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q71block": {
+ "origin": [
+ 811,
+ 1925
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q71..80"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q81block": {
+ "origin": [
+ 1125,
+ 927
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q81..90"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q91block": {
+ "origin": [
+ 1125,
+ 1258
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q91..100"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ }
+ },
+ "preProcessors": [
+ {
+ "name": "CropPage",
+ "options": {
+ "morphKernel": [
+ 10,
+ 10
+ ]
+ }
+ }
+ ]
+}
diff --git a/samples/community/UmarFarootAPS/answer_key.csv b/samples/community/UmarFarootAPS/answer_key.csv
new file mode 100644
index 00000000..b40e8959
--- /dev/null
+++ b/samples/community/UmarFarootAPS/answer_key.csv
@@ -0,0 +1,200 @@
+q1,C
+q2,C
+q3,"D,E"
+q4,"A,AB"
+q5,"[['A', '1'], ['B', '2']]"
+q6,"['A', 'B']"
+q7,C
+q8,D
+q9,B
+q10,B
+q11,A
+q12,A
+q13,C
+q14,B
+q15,D
+q16,B
+q17,C
+q18,A
+q19,B
+q20,D
+q21,D
+q22,C
+q23,A
+q24,C
+q25,D
+q26,C
+q27,C
+q28,B
+q29,A
+q30,D
+q31,C
+q32,B
+q33,B
+q34,C
+q35,A
+q36,D
+q37,C
+q38,B
+q39,C
+q40,A
+q41,A
+q42,C
+q43,D
+q44,D
+q45,B
+q46,C
+q47,C
+q48,A
+q49,C
+q50,B
+q51,B
+q52,C
+q53,D
+q54,C
+q55,B
+q56,B
+q57,A
+q58,A
+q59,D
+q60,C
+q61,C
+q62,B
+q63,A
+q64,C
+q65,D
+q66,C
+q67,B
+q68,A
+q69,B
+q70,B
+q71,C
+q72,B
+q73,C
+q74,A
+q75,A
+q76,C
+q77,D
+q78,D
+q79,B
+q80,A
+q81,B
+q82,C
+q83,D
+q84,C
+q85,A
+q86,C
+q87,D
+q88,B
+q89,C
+q90,B
+q91,B
+q92,A
+q93,C
+q94,D
+q95,C
+q96,B
+q97,B
+q98,A
+q99,A
+q100,A
+q101,A
+q102,B
+q103,C
+q104,C
+q105,A
+q106,D
+q107,B
+q108,A
+q109,C
+q110,B
+q111,B
+q112,C
+q113,C
+q114,B
+q115,D
+q116,B
+q117,A
+q118,C
+q119,D
+q120,C
+q121,C
+q122,A
+q123,B
+q124,C
+q125,D
+q126,C
+q127,C
+q128,D
+q129,D
+q130,A
+q131,A
+q132,C
+q133,B
+q134,C
+q135,D
+q136,B
+q137,C
+q138,A
+q139,B
+q140,D
+q141,D
+q142,C
+q143,D
+q144,A
+q145,A
+q146,C
+q147,A
+q148,D
+q149,D
+q150,B
+q151,A
+q152,B
+q153,B
+q154,D
+q155,D
+q156,B
+q157,A
+q158,B
+q159,A
+q160,C
+q161,D
+q162,C
+q163,A
+q164,B
+q165,D
+q166,D
+q167,C
+q168,C
+q169,C
+q170,D
+q171,A
+q172,A
+q173,C
+q174,C
+q175,B
+q176,D
+q177,A
+q178,B
+q179,B
+q180,C
+q181,D
+q182,C
+q183,B
+q184,B
+q185,C
+q186,D
+q187,D
+q188,A
+q189,A
+q190,B
+q191,C
+q192,B
+q193,D
+q194,C
+q195,B
+q196,B
+q197,A
+q198,B
+q199,B
+q200,A
diff --git a/samples/community/UmarFarootAPS/config.json b/samples/community/UmarFarootAPS/config.json
new file mode 100644
index 00000000..fde8f5b2
--- /dev/null
+++ b/samples/community/UmarFarootAPS/config.json
@@ -0,0 +1,11 @@
+{
+ "dimensions": {
+ "display_height": 960,
+ "display_width": 1280,
+ "processing_height": 1640,
+ "processing_width": 1332
+ },
+ "outputs": {
+ "show_image_level": 0
+ }
+}
diff --git a/samples/community/UmarFarootAPS/evaluation.json b/samples/community/UmarFarootAPS/evaluation.json
new file mode 100644
index 00000000..14a3db25
--- /dev/null
+++ b/samples/community/UmarFarootAPS/evaluation.json
@@ -0,0 +1,14 @@
+{
+ "source_type": "csv",
+ "options": {
+ "answer_key_csv_path": "answer_key.csv",
+ "should_explain_scoring": true
+ },
+ "marking_schemes": {
+ "DEFAULT": {
+ "correct": "1",
+ "incorrect": "0",
+ "unmarked": "0"
+ }
+ }
+}
diff --git a/samples/sample3/omr_marker.jpg b/samples/community/UmarFarootAPS/omr_marker.jpg
similarity index 100%
rename from samples/sample3/omr_marker.jpg
rename to samples/community/UmarFarootAPS/omr_marker.jpg
diff --git a/samples/community/UmarFarootAPS/scans/scan-type-1.jpg b/samples/community/UmarFarootAPS/scans/scan-type-1.jpg
new file mode 100644
index 00000000..0932b240
Binary files /dev/null and b/samples/community/UmarFarootAPS/scans/scan-type-1.jpg differ
diff --git a/samples/community/UmarFarootAPS/scans/scan-type-2.jpg b/samples/community/UmarFarootAPS/scans/scan-type-2.jpg
new file mode 100644
index 00000000..2eef7c32
Binary files /dev/null and b/samples/community/UmarFarootAPS/scans/scan-type-2.jpg differ
diff --git a/samples/community/UmarFarootAPS/template.json b/samples/community/UmarFarootAPS/template.json
new file mode 100644
index 00000000..f096f518
--- /dev/null
+++ b/samples/community/UmarFarootAPS/template.json
@@ -0,0 +1,188 @@
+{
+ "pageDimensions": [
+ 2550,
+ 3300
+ ],
+ "bubbleDimensions": [
+ 32,
+ 32
+ ],
+ "preProcessors": [
+ {
+ "name": "CropOnMarkers",
+ "options": {
+ "relativePath": "omr_marker.jpg",
+ "sheetToMarkerWidthRatio": 17
+ }
+ }
+ ],
+ "customLabels": {
+ "Roll_no": [
+ "r1",
+ "r2",
+ "r3",
+ "r4"
+ ]
+ },
+ "fieldBlocks": {
+ "Roll_no": {
+ "fieldType": "QTYPE_INT",
+ "origin": [
+ 2169,
+ 180
+ ],
+ "fieldLabels": [
+ "r1",
+ "r2",
+ "r3",
+ "r4"
+ ],
+ "bubblesGap": 61,
+ "labelsGap": 93
+ },
+ "MCQBlock1a1": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 197,
+ 300
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q1..17"
+ ]
+ },
+ "MCQBlock1a2": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 197,
+ 1310
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q18..34"
+ ]
+ },
+ "MCQBlock1a3": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 197,
+ 2316
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q35..50"
+ ]
+ },
+ "MCQBlock1a4": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 725,
+ 300
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q51..67"
+ ]
+ },
+ "MCQBlock1a5": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 725,
+ 1310
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q68..84"
+ ]
+ },
+ "MCQBlock1a6": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 725,
+ 2316
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q85..100"
+ ]
+ },
+ "MCQBlock1a7": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 1250,
+ 300
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q101..117"
+ ]
+ },
+ "MCQBlock1a8": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 1250,
+ 1310
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q118..134"
+ ]
+ },
+ "MCQBlock1a9": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 1250,
+ 2316
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q135..150"
+ ]
+ },
+ "MCQBlock1a10": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 1770,
+ 300
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q151..167"
+ ]
+ },
+ "MCQBlock1a11": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 1770,
+ 1310
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q168..184"
+ ]
+ },
+ "MCQBlock1a12": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 1770,
+ 2316
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q185..200"
+ ]
+ }
+ }
+}
diff --git a/samples/community/dxuian/omrcollegesheet.jpg b/samples/community/dxuian/omrcollegesheet.jpg
new file mode 100644
index 00000000..b9d70c6b
Binary files /dev/null and b/samples/community/dxuian/omrcollegesheet.jpg differ
diff --git a/samples/community/dxuian/template.json b/samples/community/dxuian/template.json
new file mode 100644
index 00000000..43bf2be6
--- /dev/null
+++ b/samples/community/dxuian/template.json
@@ -0,0 +1,48 @@
+{
+ "pageDimensions": [707, 484],
+ "bubbleDimensions": [15, 10],
+ "fieldBlocks": {
+ "Column1": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [82, 35],
+ "bubblesGap": 21,
+ "labelsGap": 22.7,
+ "bubbleCount": 20,
+ "fieldLabels": ["Q1", "Q2", "Q3", "Q4", "Q5", "Q6", "Q7", "Q8", "Q9", "Q10", "Q11", "Q12", "Q13", "Q14", "Q15", "Q16", "Q17", "Q18", "Q19", "Q20"]
+ },
+ "Column2": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [205, 35],
+ "bubblesGap": 21,
+ "labelsGap": 22.7,
+ "bubbleCount": 20,
+ "fieldLabels": ["Q21", "Q22", "Q23", "Q24", "Q25", "Q26", "Q27", "Q28", "Q29", "Q30", "Q31", "Q32", "Q33", "Q34", "Q35", "Q36", "Q37", "Q38", "Q39", "Q40"]
+ },
+ "Column3": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [327, 35],
+ "bubblesGap": 21,
+ "labelsGap": 22.7,
+ "bubbleCount": 20,
+ "fieldLabels": ["Q41", "Q42", "Q43", "Q44", "Q45", "Q46", "Q47", "Q48", "Q49", "Q50", "Q51", "Q52", "Q53", "Q54", "Q55", "Q56", "Q57", "Q58", "Q59", "Q60"]
+ },
+ "Column4": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [450, 35],
+ "bubblesGap": 21,
+ "labelsGap": 22.7,
+ "bubbleCount": 20,
+ "fieldLabels": ["Q61", "Q62", "Q63", "Q64", "Q65", "Q66", "Q67", "Q68", "Q69", "Q70", "Q71", "Q72", "Q73", "Q74", "Q75", "Q76", "Q77", "Q78", "Q79", "Q80"]
+ },
+ "Column5": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [573, 35],
+ "bubblesGap": 21,
+ "labelsGap": 22.7,
+ "bubbleCount": 20,
+ "fieldLabels": ["Q81", "Q82", "Q83", "Q84", "Q85", "Q86", "Q87", "Q88", "Q89", "Q90", "Q91", "Q92", "Q93", "Q94", "Q95", "Q96", "Q97", "Q98", "Q99", "Q100"]
+ }
+ },
+
+ "emptyValue": "-"
+}
\ No newline at end of file
diff --git a/samples/community/ibrahimkilic/template.json b/samples/community/ibrahimkilic/template.json
new file mode 100644
index 00000000..f0cb9cf3
--- /dev/null
+++ b/samples/community/ibrahimkilic/template.json
@@ -0,0 +1,30 @@
+{
+ "pageDimensions": [
+ 299,
+ 328
+ ],
+ "bubbleDimensions": [
+ 20,
+ 20
+ ],
+ "emptyValue": "no",
+ "fieldBlocks": {
+ "YesNoBlock1": {
+ "direction": "horizontal",
+ "bubbleValues": [
+ "yes"
+ ],
+ "origin": [
+ 15,
+ 55
+ ],
+ "emptyValue": "no",
+ "bubblesGap": 48,
+ "labelsGap": 48,
+ "fieldLabels": [
+ "q1..5"
+ ]
+ }
+ },
+ "preProcessors": []
+}
diff --git a/samples/community/ibrahimkilic/yes_no_questionnarie.jpg b/samples/community/ibrahimkilic/yes_no_questionnarie.jpg
new file mode 100644
index 00000000..e9436f41
Binary files /dev/null and b/samples/community/ibrahimkilic/yes_no_questionnarie.jpg differ
diff --git a/samples/community/samuelIkoli/template.json b/samples/community/samuelIkoli/template.json
new file mode 100644
index 00000000..759f6972
--- /dev/null
+++ b/samples/community/samuelIkoli/template.json
@@ -0,0 +1,28 @@
+{
+ "pageDimensions": [630, 404],
+ "bubbleDimensions": [20, 15],
+ "customLabels": {},
+ "fieldBlocks": {
+ "MCQBlock1": {
+ "fieldType": "QTYPE_MCQ5",
+ "origin": [33, 6],
+ "fieldLabels": ["q1", "q2", "q3", "q4", "q5", "q6", "q7", "q8", "q9", "q10", "q11", "q12", "q13", "q14", "q15", "q16", "q17", "q18", "q19", "q20"],
+ "bubblesGap": 33,
+ "labelsGap": 20
+ },
+ "MCQBlock2": {
+ "fieldType": "QTYPE_MCQ5",
+ "origin": [248, 6],
+ "fieldLabels": ["q21", "q22", "q23", "q24", "q25", "q26", "q27", "q28", "q29", "q30", "q31", "q32", "q33", "q34", "q35", "q36", "q37", "q38", "q39", "q40"],
+ "bubblesGap": 33,
+ "labelsGap": 20
+ },
+ "MCQBlock3": {
+ "fieldType": "QTYPE_MCQ5",
+ "origin": [465, 6],
+ "fieldLabels": ["q41", "q42", "q43", "q44", "q45", "q46", "q47", "q48", "q49", "q50", "q51", "q52", "q53", "q54", "q55", "q56", "q57", "q58", "q59", "q60"],
+ "bubblesGap": 33,
+ "labelsGap": 20
+ }
+ }
+}
diff --git a/samples/community/samuelIkoli/waec_sample.jpeg b/samples/community/samuelIkoli/waec_sample.jpeg
new file mode 100644
index 00000000..fcd61e80
Binary files /dev/null and b/samples/community/samuelIkoli/waec_sample.jpeg differ
diff --git a/samples/sample1/config.json b/samples/sample1/config.json
new file mode 100644
index 00000000..a9daf449
--- /dev/null
+++ b/samples/sample1/config.json
@@ -0,0 +1,11 @@
+{
+ "dimensions": {
+ "display_height": 2480,
+ "display_width": 1640,
+ "processing_height": 820,
+ "processing_width": 666
+ },
+ "outputs": {
+ "show_image_level": 5
+ }
+}
diff --git a/samples/sample1/template.json b/samples/sample1/template.json
index ec0f9c68..46635774 100644
--- a/samples/sample1/template.json
+++ b/samples/sample1/template.json
@@ -1,260 +1,197 @@
{
- "Dimensions": [
+ "pageDimensions": [
1846,
1500
],
- "BubbleDimensions": [
+ "bubbleDimensions": [
40,
40
],
- "Options": {
- "Marker": {
- "RelativePath": "omr_marker.jpg",
- "SheetToMarkerWidthRatio": 17
- }
- },
- "Concatenations": {
+ "customLabels": {
"Roll": [
- "Squad",
"Medium",
- "roll0",
- "roll1",
- "roll2",
- "roll3",
- "roll4",
- "roll5",
- "roll6",
- "roll7",
- "roll8"
+ "roll1..9"
],
"q5": [
- "q5.1",
- "q5.2"
+ "q5_1",
+ "q5_2"
],
"q6": [
- "q6.1",
- "q6.2"
+ "q6_1",
+ "q6_2"
],
"q7": [
- "q7.1",
- "q7.2"
+ "q7_1",
+ "q7_2"
],
- "q9": [
- "q9.1",
- "q9.2"
+ "q8": [
+ "q8_1",
+ "q8_2"
],
- "q11": [
- "q8.1",
- "q8.2"
+ "q9": [
+ "q9_1",
+ "q9_2"
]
},
- "Singles": [
- "q1",
- "q2",
- "q3",
- "q4",
- "q10",
- "q11",
- "q12",
- "q13",
- "q14",
- "q15",
- "q16",
- "q17",
- "q18",
- "q19",
- "q20"
- ],
- "QBlocks": {
+ "fieldBlocks": {
"Medium": {
- "qType": "QTYPE_MED",
- "orig": [
- 160,
- 285
- ],
- "bigGaps": [
- 115,
- 11
- ],
- "gaps": [
- 59,
- 46
- ],
- "qNos": [
- [
- [
- "Medium"
- ]
- ]
+ "bubblesGap": 41,
+ "bubbleValues": [
+ "E",
+ "H"
+ ],
+ "direction": "vertical",
+ "fieldLabels": [
+ "Medium"
+ ],
+ "labelsGap": 0,
+ "origin": [
+ 170,
+ 282
]
},
"Roll": {
- "qType": "QTYPE_ROLL",
- "orig": [
- 218,
- 285
- ],
- "bigGaps": [
- 115,
- 11
- ],
- "gaps": [
- 58,
- 46
- ],
- "qNos": [
- [
- [
- "roll0",
- "roll1",
- "roll2",
- "roll3",
- "roll4",
- "roll5",
- "roll6",
- "roll7",
- "roll8"
- ]
- ]
+ "fieldType": "QTYPE_INT",
+ "fieldLabels": [
+ "roll1..9"
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 58,
+ "origin": [
+ 225,
+ 282
]
},
- "Int1": {
- "qType": "QTYPE_INT",
- "orig": [
+ "Int_Block_Q5": {
+ "fieldType": "QTYPE_INT",
+ "fieldLabels": [
+ "q5_1",
+ "q5_2"
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 60,
+ "origin": [
903,
- 285
- ],
- "bigGaps": [
- 128,
- 11
- ],
- "gaps": [
- 59,
- 46
- ],
- "qNos": [
- [
- [
- "q5.1",
- "q5.2"
- ],
- [
- "q6.1",
- "q6.2"
- ],
- [
- "q7.1",
- "q7.2"
- ]
- ]
+ 282
]
},
- "Int2": {
- "qType": "QTYPE_INT",
- "orig": [
- 1418,
- 285
- ],
- "bigGaps": [
- 128,
- 11
- ],
- "gaps": [
- 59,
- 46
- ],
- "qNos": [
- [
- [
- "q8.1",
- "q8.2"
- ],
- [
- "q9.1",
- "q9.2"
- ]
- ]
+ "Int_Block_Q6": {
+ "fieldType": "QTYPE_INT",
+ "fieldLabels": [
+ "q6_1",
+ "q6_2"
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 60,
+ "origin": [
+ 1077,
+ 282
]
},
- "Mcq1": {
- "qType": "QTYPE_MCQ4",
- "orig": [
- 118,
- 857
- ],
- "bigGaps": [
- 115,
- 181
- ],
- "gaps": [
- 59,
- 53
- ],
- "qNos": [
- [
- [
- "q1",
- "q2",
- "q3",
- "q4"
- ],
- [
- "q10",
- "q11",
- "q12",
- "q13"
- ]
- ]
+ "Int_Block_Q7": {
+ "fieldType": "QTYPE_INT",
+ "fieldLabels": [
+ "q7_1",
+ "q7_2"
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 60,
+ "origin": [
+ 1240,
+ 282
+ ]
+ },
+ "Int_Block_Q8": {
+ "fieldType": "QTYPE_INT",
+ "fieldLabels": [
+ "q8_1",
+ "q8_2"
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 57,
+ "origin": [
+ 1410,
+ 282
+ ]
+ },
+ "Int_Block_Q9": {
+ "fieldType": "QTYPE_INT",
+ "fieldLabels": [
+ "q9_1",
+ "q9_2"
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 57,
+ "origin": [
+ 1580,
+ 282
]
},
- "Mcq2": {
- "qType": "QTYPE_MCQ4",
- "orig": [
+ "MCQ_Block_Q1": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q1..4"
+ ],
+ "bubblesGap": 59,
+ "labelsGap": 50,
+ "origin": [
+ 121,
+ 860
+ ]
+ },
+ "MCQ_Block_Q10": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q10..13"
+ ],
+ "bubblesGap": 59,
+ "labelsGap": 50,
+ "origin": [
+ 121,
+ 1195
+ ]
+ },
+ "MCQ_Block_Q14": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q14..16"
+ ],
+ "bubblesGap": 57,
+ "labelsGap": 50,
+ "origin": [
905,
860
- ],
- "bigGaps": [
- 115,
- 180
- ],
- "gaps": [
- 59,
- 53
- ],
- "qNos": [
- [
- [
- "q14",
- "q15",
- "q16"
- ]
- ]
]
},
- "Mcq3": {
- "qType": "QTYPE_MCQ4",
- "orig": [
+ "MCQ_Block_Q17": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q17..20"
+ ],
+ "bubblesGap": 57,
+ "labelsGap": 50,
+ "origin": [
905,
- 1198
- ],
- "bigGaps": [
- 115,
- 180
- ],
- "gaps": [
- 59,
- 53
- ],
- "qNos": [
- [
- [
- "q17",
- "q18",
- "q19",
- "q20"
- ]
- ]
+ 1195
]
}
- }
-}
\ No newline at end of file
+ },
+ "preProcessors": [
+ {
+ "name": "CropPage",
+ "options": {
+ "morphKernel": [
+ 10,
+ 10
+ ]
+ }
+ },
+ {
+ "name": "CropOnMarkers",
+ "options": {
+ "relativePath": "omr_marker.jpg",
+ "sheetToMarkerWidthRatio": 17
+ }
+ }
+ ]
+}
diff --git a/samples/sample2/AdrianSample/adrian_omr.png b/samples/sample2/AdrianSample/adrian_omr.png
index dc008145..69a53823 100644
Binary files a/samples/sample2/AdrianSample/adrian_omr.png and b/samples/sample2/AdrianSample/adrian_omr.png differ
diff --git a/samples/sample2/AdrianSample/adrian_omr_2.png b/samples/sample2/AdrianSample/adrian_omr_2.png
new file mode 100644
index 00000000..d8db0994
Binary files /dev/null and b/samples/sample2/AdrianSample/adrian_omr_2.png differ
diff --git a/samples/sample2/config.json b/samples/sample2/config.json
new file mode 100644
index 00000000..a9daf449
--- /dev/null
+++ b/samples/sample2/config.json
@@ -0,0 +1,11 @@
+{
+ "dimensions": {
+ "display_height": 2480,
+ "display_width": 1640,
+ "processing_height": 820,
+ "processing_width": 666
+ },
+ "outputs": {
+ "show_image_level": 5
+ }
+}
diff --git a/samples/sample2/template.json b/samples/sample2/template.json
index e82a9051..41ec9ffa 100644
--- a/samples/sample2/template.json
+++ b/samples/sample2/template.json
@@ -1,46 +1,35 @@
{
- "Dimensions": [
+ "pageDimensions": [
300,
400
],
- "BubbleDimensions": [
+ "bubbleDimensions": [
25,
25
],
- "Concatenations": {},
- "Singles": [
- "q1",
- "q2",
- "q3",
- "q4",
- "q5"
+ "preProcessors": [
+ {
+ "name": "CropPage",
+ "options": {
+ "morphKernel": [
+ 10,
+ 10
+ ]
+ }
+ }
],
- "QBlocks": {
- "MCQBlock1": {
- "qType": "QTYPE_MCQ5",
- "orig": [
+ "fieldBlocks": {
+ "MCQ_Block_1": {
+ "fieldType": "QTYPE_MCQ5",
+ "origin": [
65,
60
],
- "qNos": [
- [
- [
- "q1",
- "q2",
- "q3",
- "q4",
- "q5"
- ]
- ]
- ],
- "gaps": [
- 41,
- 52
+ "fieldLabels": [
+ "q1..5"
],
- "bigGaps": [
- 30,
- 30
- ]
+ "labelsGap": 52,
+ "bubblesGap": 41
}
}
-}
\ No newline at end of file
+}
diff --git a/samples/sample3/README.md b/samples/sample3/README.md
new file mode 100644
index 00000000..d764c8b3
--- /dev/null
+++ b/samples/sample3/README.md
@@ -0,0 +1,31 @@
+## Observation
+The OMR layout is slightly different on colored thick papers vs on xeroxed thin papers. The shift becomes noticible in case of OMR with large number of questions.
+
+We overlapped a colored OMR sheet with a xerox copy of the same OMR sheet(both printed on A4 papers) and observed that there is a great amount of layout sheet as we reach the bottom of the OMR.
+
+Link to an explainer with a real life example: [Google Drive](https://drive.google.com/drive/folders/1GpZTmpEhEjSALJEMjHwDafzWKgEoCTOI?usp=sharing)
+
+## Reasons for shifts in Template layout:
+Listing out a few reasons for the above observation:
+### Printer margin setting
+The margin settings for different printers may be different for the same OMR layout. Thus causing the print to become elongated either horizontally, vertically or both ways.
+
+### The Fan-out effect
+The fan-out effect is usually observed in a sheet fed offset press. Depending on how the papers are made, their dimensions have a tendency to change when they are exposed to moisture or water.
+
+The standard office papers(80 gsm) can easily capture moisture and change shape e.g. in case they get stored for multiple days in a place where the weather is highly humid.
+
+Below are some examples of the GSM ranges:
+
+- 74gsm to 90gsm – This is the basic standard office paper, used in your laser printers.
+- 100gsm to 120gsm – This is stationary paper used for standard letterheads, complimentary slips.
+- 130 to 170gsm – Mostly used for leaflets, posters, single-sided flyers, and brochures.
+
+## Solution
+
+It is recommended to scan each types of prints into different folders and use a separate template.json layout for each of the folders. The same is presented in this sample folder.
+
+## References
+
+- [Paper dimensional stability in sheet-fed offset printing](https://www.diva-portal.org/smash/get/diva2:517895/FULLTEXT01.pdf)
+- An analysis of a few ["Interesting" bubble sheets](https://imgur.com/a/10qwL)
diff --git a/samples/sample3/colored-thick-sheet/rgb-100-gsm.jpg b/samples/sample3/colored-thick-sheet/rgb-100-gsm.jpg
new file mode 100644
index 00000000..4a34731a
Binary files /dev/null and b/samples/sample3/colored-thick-sheet/rgb-100-gsm.jpg differ
diff --git a/samples/sample3/colored-thick-sheet/template.json b/samples/sample3/colored-thick-sheet/template.json
new file mode 100644
index 00000000..743723d6
--- /dev/null
+++ b/samples/sample3/colored-thick-sheet/template.json
@@ -0,0 +1,143 @@
+{
+ "pageDimensions": [
+ 1800,
+ 2400
+ ],
+ "bubbleDimensions": [
+ 23,
+ 20
+ ],
+ "fieldBlocks": {
+ "q01block": {
+ "origin": [
+ 504,
+ 927
+ ],
+ "bubblesGap": 60.35,
+ "labelsGap": 31.75,
+ "fieldLabels": [
+ "q1..10"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q11block": {
+ "origin": [
+ 504,
+ 1242
+ ],
+ "bubblesGap": 60.35,
+ "labelsGap": 31.75,
+ "fieldLabels": [
+ "q11..20"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q21block": {
+ "origin": [
+ 500,
+ 1562
+ ],
+ "bubblesGap": 61.25,
+ "labelsGap": 32.5,
+ "fieldLabels": [
+ "q21..30"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q31block": {
+ "origin": [
+ 500,
+ 1885
+ ],
+ "bubblesGap": 62.25,
+ "labelsGap": 33.5,
+ "fieldLabels": [
+ "q31..40"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q41block": {
+ "origin": [
+ 811,
+ 927
+ ],
+ "bubblesGap": 60.35,
+ "labelsGap": 31.75,
+ "fieldLabels": [
+ "q41..50"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q51block": {
+ "origin": [
+ 811,
+ 1242
+ ],
+ "bubblesGap": 60.35,
+ "labelsGap": 31.75,
+ "fieldLabels": [
+ "q51..60"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q61block": {
+ "origin": [
+ 811,
+ 1562
+ ],
+ "bubblesGap": 61.25,
+ "labelsGap": 32.5,
+ "fieldLabels": [
+ "q61..70"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q71block": {
+ "origin": [
+ 811,
+ 1885
+ ],
+ "bubblesGap": 62.25,
+ "labelsGap": 33.5,
+ "fieldLabels": [
+ "q71..80"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q81block": {
+ "origin": [
+ 1120,
+ 927
+ ],
+ "bubblesGap": 60.35,
+ "labelsGap": 31.75,
+ "fieldLabels": [
+ "q81..90"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q91block": {
+ "origin": [
+ 1120,
+ 1242
+ ],
+ "bubblesGap": 60.35,
+ "labelsGap": 31.75,
+ "fieldLabels": [
+ "q91..100"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ }
+ },
+ "preProcessors": [
+ {
+ "name": "CropPage",
+ "options": {
+ "morphKernel": [
+ 10,
+ 10
+ ]
+ }
+ }
+ ]
+}
diff --git a/samples/sample3/config.json b/samples/sample3/config.json
new file mode 100644
index 00000000..a9daf449
--- /dev/null
+++ b/samples/sample3/config.json
@@ -0,0 +1,11 @@
+{
+ "dimensions": {
+ "display_height": 2480,
+ "display_width": 1640,
+ "processing_height": 820,
+ "processing_width": 666
+ },
+ "outputs": {
+ "show_image_level": 5
+ }
+}
diff --git a/samples/sample3/template.json b/samples/sample3/template.json
deleted file mode 100644
index 2dddf37e..00000000
--- a/samples/sample3/template.json
+++ /dev/null
@@ -1,248 +0,0 @@
-{
- "Dimensions": [
- 1846,
- 1500
- ],
- "BubbleDimensions": [
- 40,
- 40
- ],
- "Options": {
- "Marker": {
- "RelativePath": "omr_marker.jpg",
- "SheetToMarkerWidthRatio": 17
- },
- "OverrideFlags":{
- "noCropping": true
- }
- },
- "Concatenations": {
- "Roll": [
- "Squad",
- "Medium",
- "roll0",
- "roll1",
- "roll2",
- "roll3",
- "roll4",
- "roll5",
- "roll6",
- "roll7",
- "roll8"
- ],
- "q6": [
- "q6.1",
- "q6.2"
- ],
- "q7": [
- "q7.1",
- "q7.2"
- ],
- "q8": [
- "q8.1",
- "q8.2"
- ],
- "q9": [
- "q9.1",
- "q9.2"
- ],
- "q10": [
- "q10.1",
- "q10.2"
- ]
- },
- "Singles": [
- "q1",
- "q2",
- "q3",
- "q4",
- "q5",
- "q11",
- "q12",
- "q13",
- "q14",
- "q15",
- "q16",
- "q17",
- "q18",
- "q19",
- "q20",
- "q21",
- "q22"
- ],
- "QBlocks": {
- "Medium": {
- "qType": "QTYPE_MED",
- "orig": [
- 208,
- 205
- ],
- "bigGaps": [
- 115,
- 11
- ],
- "gaps": [
- 59,
- 46
- ],
- "qNos": [
- [
- [
- "Medium"
- ]
- ]
- ]
- },
- "Roll": {
- "qType": "QTYPE_ROLL",
- "orig": [
- 261,
- 210
- ],
- "bigGaps": [
- 115,
- 11
- ],
- "gaps": [
- 58,
- 46
- ],
- "qNos": [
- [
- [
- "roll0",
- "roll1",
- "roll2",
- "roll3",
- "roll4",
- "roll5",
- "roll6",
- "roll7",
- "roll8"
- ]
- ]
- ]
- },
- "Int1": {
- "qType": "QTYPE_INT",
- "orig": [
- 935,
- 211
- ],
- "bigGaps": [
- 124,
- 11
- ],
- "gaps": [
- 57,
- 46
- ],
- "qNos": [
- [
- [
- "q6.1",
- "q6.2"
- ],
- [
- "q7.1",
- "q7.2"
- ],
- [
- "q8.1",
- "q8.2"
- ],
- [
- "q9.1",
- "q9.2"
- ],
- [
- "q10.1",
- "q10.2"
- ]
- ]
- ]
- },
- "Mcq1": {
- "qType": "QTYPE_MCQ4",
- "orig": [
- 198,
- 826
- ],
- "bigGaps": [
- 115,
- 183
- ],
- "gaps": [
- 93,
- 62
- ],
- "qNos": [
- [
- [
- "q1",
- "q2",
- "q3",
- "q4",
- "q5"
- ]
- ]
- ]
- },
- "Mcq2": {
- "qType": "QTYPE_MCQ4",
- "orig": [
- 833,
- 830
- ],
- "bigGaps": [
- 127,
- 254
- ],
- "gaps": [
- 71,
- 61
- ],
- "qNos": [
- [
- [
- "q11",
- "q12",
- "q13",
- "q14"
- ],
- [
- "q15",
- "q16",
- "q17",
- "q18"
- ]
- ]
- ]
- },
- "Mcq3": {
- "qType": "QTYPE_MCQ4",
- "orig": [
- 1481,
- 830
- ],
- "bigGaps": [
- 115,
- 183
- ],
- "gaps": [
- 73,
- 61
- ],
- "qNos": [
- [
- [
- "q19",
- "q20",
- "q21",
- "q22"
- ]
- ]
- ]
- }
- }
-}
\ No newline at end of file
diff --git a/samples/sample3/xeroxed-thin-sheet/grayscale-80-gsm.jpg b/samples/sample3/xeroxed-thin-sheet/grayscale-80-gsm.jpg
new file mode 100644
index 00000000..3e0e3efa
Binary files /dev/null and b/samples/sample3/xeroxed-thin-sheet/grayscale-80-gsm.jpg differ
diff --git a/samples/sample3/xeroxed-thin-sheet/template.json b/samples/sample3/xeroxed-thin-sheet/template.json
new file mode 100644
index 00000000..8aa4e41c
--- /dev/null
+++ b/samples/sample3/xeroxed-thin-sheet/template.json
@@ -0,0 +1,143 @@
+{
+ "pageDimensions": [
+ 1800,
+ 2400
+ ],
+ "bubbleDimensions": [
+ 23,
+ 20
+ ],
+ "fieldBlocks": {
+ "q01block": {
+ "origin": [
+ 492,
+ 924
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q1..10"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q11block": {
+ "origin": [
+ 492,
+ 1258
+ ],
+ "bubblesGap": 59.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q11..20"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q21block": {
+ "origin": [
+ 492,
+ 1589
+ ],
+ "bubblesGap": 60.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q21..30"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q31block": {
+ "origin": [
+ 487,
+ 1920
+ ],
+ "bubblesGap": 61.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q31..40"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q41block": {
+ "origin": [
+ 807,
+ 924
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q41..50"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q51block": {
+ "origin": [
+ 803,
+ 1258
+ ],
+ "bubblesGap": 59.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q51..60"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q61block": {
+ "origin": [
+ 803,
+ 1589
+ ],
+ "bubblesGap": 60.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q61..70"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q71block": {
+ "origin": [
+ 803,
+ 1920
+ ],
+ "bubblesGap": 60.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q71..80"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q81block": {
+ "origin": [
+ 1115,
+ 924
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q81..90"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q91block": {
+ "origin": [
+ 1115,
+ 1258
+ ],
+ "bubblesGap": 59.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q91..100"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ }
+ },
+ "preProcessors": [
+ {
+ "name": "CropPage",
+ "options": {
+ "morphKernel": [
+ 10,
+ 10
+ ]
+ }
+ }
+ ]
+}
diff --git a/samples/sample4/Class1/sheet1.jpg b/samples/sample4/Class1/sheet1.jpg
deleted file mode 100644
index f3248903..00000000
Binary files a/samples/sample4/Class1/sheet1.jpg and /dev/null differ
diff --git a/samples/sample4/Class2/sheet1.jpg b/samples/sample4/Class2/sheet1.jpg
deleted file mode 100644
index f3248903..00000000
Binary files a/samples/sample4/Class2/sheet1.jpg and /dev/null differ
diff --git a/samples/sample4/IMG_20201116_143512.jpg b/samples/sample4/IMG_20201116_143512.jpg
new file mode 100644
index 00000000..2f591c7a
Binary files /dev/null and b/samples/sample4/IMG_20201116_143512.jpg differ
diff --git a/samples/sample4/IMG_20201116_150717658.jpg b/samples/sample4/IMG_20201116_150717658.jpg
new file mode 100644
index 00000000..592fe425
Binary files /dev/null and b/samples/sample4/IMG_20201116_150717658.jpg differ
diff --git a/samples/sample4/IMG_20201116_150750830.jpg b/samples/sample4/IMG_20201116_150750830.jpg
new file mode 100644
index 00000000..6846ef31
Binary files /dev/null and b/samples/sample4/IMG_20201116_150750830.jpg differ
diff --git a/samples/sample4/config.json b/samples/sample4/config.json
new file mode 100644
index 00000000..df59a460
--- /dev/null
+++ b/samples/sample4/config.json
@@ -0,0 +1,13 @@
+{
+ "dimensions": {
+ "display_width": 1189,
+ "display_height": 1682
+ },
+ "threshold_params": {
+ "MIN_JUMP": 30
+ },
+ "outputs": {
+ "filter_out_multimarked_files": false,
+ "show_image_level": 5
+ }
+}
diff --git a/samples/sample4/evaluation.json b/samples/sample4/evaluation.json
new file mode 100644
index 00000000..50db0fbe
--- /dev/null
+++ b/samples/sample4/evaluation.json
@@ -0,0 +1,34 @@
+{
+ "source_type": "custom",
+ "options": {
+ "questions_in_order": [
+ "q1..11"
+ ],
+ "answers_in_order": [
+ "B",
+ "D",
+ "C",
+ "B",
+ "D",
+ "C",
+ [
+ "B",
+ "C",
+ "BC"
+ ],
+ "A",
+ "C",
+ "D",
+ "C"
+ ],
+ "should_explain_scoring": true,
+ "enable_evaluation_table_to_csv": true
+ },
+ "marking_schemes": {
+ "DEFAULT": {
+ "correct": "3",
+ "incorrect": "-1",
+ "unmarked": "0"
+ }
+ }
+}
diff --git a/samples/sample4/omr_marker.jpg b/samples/sample4/omr_marker.jpg
deleted file mode 100644
index 201dab0a..00000000
Binary files a/samples/sample4/omr_marker.jpg and /dev/null differ
diff --git a/samples/sample4/template.json b/samples/sample4/template.json
index 958d6ef8..0d615b87 100644
--- a/samples/sample4/template.json
+++ b/samples/sample4/template.json
@@ -1,263 +1,45 @@
{
- "Dimensions": [
- 1846,
- 1500
+ "pageDimensions": [
+ 1189,
+ 1682
],
- "BubbleDimensions": [
- 40,
- 40
+ "bubbleDimensions": [
+ 30,
+ 30
],
- "Options": {
- "Marker": {
- "RelativePath": "omr_marker.jpg",
- "SheetToMarkerWidthRatio": 17
+ "preProcessors": [
+ {
+ "name": "GaussianBlur",
+ "options": {
+ "kSize": [
+ 3,
+ 3
+ ],
+ "sigmaX": 0
+ }
},
- "OverrideFlags": {
- "noCropping": true
+ {
+ "name": "CropPage",
+ "options": {
+ "morphKernel": [
+ 10,
+ 10
+ ]
+ }
}
- },
- "Concatenations": {
- "Roll": [
- "Squad",
- "Medium",
- "roll0",
- "roll1",
- "roll2",
- "roll3",
- "roll4",
- "roll5",
- "roll6",
- "roll7",
- "roll8"
- ],
- "q5": [
- "q5.1",
- "q5.2"
- ],
- "q6": [
- "q6.1",
- "q6.2"
- ],
- "q7": [
- "q7.1",
- "q7.2"
- ],
- "q9": [
- "q9.1",
- "q9.2"
- ],
- "q11": [
- "q8.1",
- "q8.2"
- ]
- },
- "Singles": [
- "q1",
- "q2",
- "q3",
- "q4",
- "q10",
- "q11",
- "q12",
- "q13",
- "q14",
- "q15",
- "q16",
- "q17",
- "q18",
- "q19",
- "q20"
],
- "QBlocks": {
- "Medium": {
- "qType": "QTYPE_MED",
- "orig": [
- 160,
- 285
- ],
- "bigGaps": [
- 115,
- 11
- ],
- "gaps": [
- 59,
- 46
- ],
- "qNos": [
- [
- [
- "Medium"
- ]
- ]
- ]
- },
- "Roll": {
- "qType": "QTYPE_ROLL",
- "orig": [
- 218,
- 285
- ],
- "bigGaps": [
- 115,
- 11
- ],
- "gaps": [
- 58,
- 46
- ],
- "qNos": [
- [
- [
- "roll0",
- "roll1",
- "roll2",
- "roll3",
- "roll4",
- "roll5",
- "roll6",
- "roll7",
- "roll8"
- ]
- ]
- ]
- },
- "Int1": {
- "qType": "QTYPE_INT",
- "orig": [
- 903,
- 285
- ],
- "bigGaps": [
- 128,
- 11
- ],
- "gaps": [
- 59,
- 46
- ],
- "qNos": [
- [
- [
- "q5.1",
- "q5.2"
- ],
- [
- "q6.1",
- "q6.2"
- ],
- [
- "q7.1",
- "q7.2"
- ]
- ]
- ]
- },
- "Int2": {
- "qType": "QTYPE_INT",
- "orig": [
- 1418,
- 285
- ],
- "bigGaps": [
- 128,
- 11
- ],
- "gaps": [
- 59,
- 46
- ],
- "qNos": [
- [
- [
- "q8.1",
- "q8.2"
- ],
- [
- "q9.1",
- "q9.2"
- ]
- ]
- ]
- },
- "Mcq1": {
- "qType": "QTYPE_MCQ4",
- "orig": [
- 118,
- 857
- ],
- "bigGaps": [
- 115,
- 181
- ],
- "gaps": [
- 59,
- 53
- ],
- "qNos": [
- [
- [
- "q1",
- "q2",
- "q3",
- "q4"
- ],
- [
- "q10",
- "q11",
- "q12",
- "q13"
- ]
- ]
- ]
- },
- "Mcq2": {
- "qType": "QTYPE_MCQ4",
- "orig": [
- 905,
- 860
- ],
- "bigGaps": [
- 115,
- 180
- ],
- "gaps": [
- 59,
- 53
- ],
- "qNos": [
- [
- [
- "q14",
- "q15",
- "q16"
- ]
- ]
- ]
- },
- "Mcq3": {
- "qType": "QTYPE_MCQ4",
- "orig": [
- 905,
- 1198
- ],
- "bigGaps": [
- 115,
- 180
- ],
- "gaps": [
- 59,
- 53
- ],
- "qNos": [
- [
- [
- "q17",
- "q18",
- "q19",
- "q20"
- ]
- ]
- ]
+ "fieldBlocks": {
+ "MCQBlock1": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 134,
+ 684
+ ],
+ "fieldLabels": [
+ "q1..11"
+ ],
+ "bubblesGap": 79,
+ "labelsGap": 62
}
}
-}
\ No newline at end of file
+}
diff --git a/samples/sample5/README.md b/samples/sample5/README.md
new file mode 100644
index 00000000..b5ed8ccb
--- /dev/null
+++ b/samples/sample5/README.md
@@ -0,0 +1,6 @@
+### OMRChecker Sample
+
+This sample demonstrates multiple things, namely -
+- Running OMRChecker on images scanned using popular document scanning apps
+- Using a common template.json file for sub-folders (e.g. multiple scan batches)
+- Using evaluation.json file with custom marking (without streak-based marking)
diff --git a/samples/sample3/CamScanner/sheet2.jpg b/samples/sample5/ScanBatch1/camscanner-1.jpg
similarity index 100%
rename from samples/sample3/CamScanner/sheet2.jpg
rename to samples/sample5/ScanBatch1/camscanner-1.jpg
diff --git a/samples/sample3/CamScanner/sheet1.jpg b/samples/sample5/ScanBatch2/camscanner-2.jpg
similarity index 100%
rename from samples/sample3/CamScanner/sheet1.jpg
rename to samples/sample5/ScanBatch2/camscanner-2.jpg
diff --git a/samples/sample5/config.json b/samples/sample5/config.json
new file mode 100644
index 00000000..a9daf449
--- /dev/null
+++ b/samples/sample5/config.json
@@ -0,0 +1,11 @@
+{
+ "dimensions": {
+ "display_height": 2480,
+ "display_width": 1640,
+ "processing_height": 820,
+ "processing_width": 666
+ },
+ "outputs": {
+ "show_image_level": 5
+ }
+}
diff --git a/samples/sample5/evaluation.json b/samples/sample5/evaluation.json
new file mode 100644
index 00000000..3332fe6a
--- /dev/null
+++ b/samples/sample5/evaluation.json
@@ -0,0 +1,93 @@
+{
+ "source_type": "custom",
+ "options": {
+ "questions_in_order": [
+ "q1..22"
+ ],
+ "answers_in_order": [
+ "C",
+ "C",
+ "B",
+ "C",
+ "C",
+ [
+ "1",
+ "01"
+ ],
+ "19",
+ "10",
+ "10",
+ "18",
+ "D",
+ "A",
+ "D",
+ "D",
+ "D",
+ "C",
+ "C",
+ "C",
+ "C",
+ "D",
+ "B",
+ "A"
+ ],
+ "should_explain_scoring": true
+ },
+ "marking_schemes": {
+ "DEFAULT": {
+ "correct": "1",
+ "incorrect": "0",
+ "unmarked": "0"
+ },
+ "BOOMERANG_1": {
+ "questions": [
+ "q1..5"
+ ],
+ "marking": {
+ "correct": 4,
+ "incorrect": -1,
+ "unmarked": 0
+ }
+ },
+ "PROXIMITY_1": {
+ "questions": [
+ "q6..10"
+ ],
+ "marking": {
+ "correct": 3,
+ "incorrect": -1,
+ "unmarked": 0
+ }
+ },
+ "FIBONACCI_SECTION_1": {
+ "questions": [
+ "q11..14"
+ ],
+ "marking": {
+ "correct": 2,
+ "incorrect": -1,
+ "unmarked": 0
+ }
+ },
+ "POWER_SECTION_1": {
+ "questions": [
+ "q15..18"
+ ],
+ "marking": {
+ "correct": 1,
+ "incorrect": 0,
+ "unmarked": 0
+ }
+ },
+ "FIBONACCI_SECTION_2": {
+ "questions": [
+ "q19..22"
+ ],
+ "marking": {
+ "correct": 2,
+ "incorrect": -1,
+ "unmarked": 0
+ }
+ }
+ }
+}
diff --git a/samples/sample5/omr_marker.jpg b/samples/sample5/omr_marker.jpg
new file mode 100644
index 00000000..0929feec
Binary files /dev/null and b/samples/sample5/omr_marker.jpg differ
diff --git a/samples/sample5/template.json b/samples/sample5/template.json
new file mode 100644
index 00000000..39f6e83d
--- /dev/null
+++ b/samples/sample5/template.json
@@ -0,0 +1,188 @@
+{
+ "pageDimensions": [
+ 1846,
+ 1500
+ ],
+ "bubbleDimensions": [
+ 40,
+ 40
+ ],
+ "preProcessors": [
+ {
+ "name": "CropOnMarkers",
+ "options": {
+ "relativePath": "omr_marker.jpg",
+ "sheetToMarkerWidthRatio": 17
+ }
+ }
+ ],
+ "customLabels": {
+ "Roll": [
+ "Medium",
+ "roll1..9"
+ ],
+ "q6": [
+ "q6_1",
+ "q6_2"
+ ],
+ "q7": [
+ "q7_1",
+ "q7_2"
+ ],
+ "q8": [
+ "q8_1",
+ "q8_2"
+ ],
+ "q9": [
+ "q9_1",
+ "q9_2"
+ ],
+ "q10": [
+ "q10_1",
+ "q10_2"
+ ]
+ },
+ "fieldBlocks": {
+ "Medium": {
+ "bubbleValues": [
+ "E",
+ "H"
+ ],
+ "direction": "vertical",
+ "origin": [
+ 200,
+ 215
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 0,
+ "fieldLabels": [
+ "Medium"
+ ]
+ },
+ "Roll": {
+ "fieldType": "QTYPE_INT",
+ "origin": [
+ 261,
+ 210
+ ],
+ "bubblesGap": 46.5,
+ "labelsGap": 58,
+ "fieldLabels": [
+ "roll1..9"
+ ]
+ },
+ "Int1": {
+ "fieldType": "QTYPE_INT",
+ "origin": [
+ 935,
+ 211
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 57,
+ "fieldLabels": [
+ "q6_1",
+ "q6_2"
+ ]
+ },
+ "Int2": {
+ "fieldType": "QTYPE_INT",
+ "origin": [
+ 1100,
+ 211
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 57,
+ "fieldLabels": [
+ "q7_1",
+ "q7_2"
+ ]
+ },
+ "Int3": {
+ "fieldType": "QTYPE_INT",
+ "origin": [
+ 1275,
+ 211
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 57,
+ "fieldLabels": [
+ "q8_1",
+ "q8_2"
+ ]
+ },
+ "Int4": {
+ "fieldType": "QTYPE_INT",
+ "origin": [
+ 1449,
+ 211
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 57,
+ "fieldLabels": [
+ "q9_1",
+ "q9_2"
+ ]
+ },
+ "Int5": {
+ "fieldType": "QTYPE_INT",
+ "origin": [
+ 1620,
+ 211
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 57,
+ "fieldLabels": [
+ "q10_1",
+ "q10_2"
+ ]
+ },
+ "Mcq1": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 198,
+ 826
+ ],
+ "bubblesGap": 93,
+ "labelsGap": 62,
+ "fieldLabels": [
+ "q1..5"
+ ]
+ },
+ "Mcq2": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 833,
+ 830
+ ],
+ "bubblesGap": 71,
+ "labelsGap": 61,
+ "fieldLabels": [
+ "q11..14"
+ ]
+ },
+ "Mcq3": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 833,
+ 1270
+ ],
+ "bubblesGap": 71,
+ "labelsGap": 61,
+ "fieldLabels": [
+ "q15..18"
+ ]
+ },
+ "Mcq4": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 1481,
+ 830
+ ],
+ "bubblesGap": 73,
+ "labelsGap": 61,
+ "fieldLabels": [
+ "q19..22"
+ ]
+ }
+ }
+}
diff --git a/samples/sample6/config.json b/samples/sample6/config.json
new file mode 100644
index 00000000..6f39f0ca
--- /dev/null
+++ b/samples/sample6/config.json
@@ -0,0 +1,11 @@
+{
+ "dimensions": {
+ "display_width": 2480,
+ "display_height": 3508,
+ "processing_width": 1653,
+ "processing_height": 2339
+ },
+ "outputs": {
+ "show_image_level": 5
+ }
+}
diff --git a/samples/sample6/doc-scans/sample_roll_01.jpg b/samples/sample6/doc-scans/sample_roll_01.jpg
new file mode 100644
index 00000000..25093a37
Binary files /dev/null and b/samples/sample6/doc-scans/sample_roll_01.jpg differ
diff --git a/samples/sample6/doc-scans/sample_roll_02.jpg b/samples/sample6/doc-scans/sample_roll_02.jpg
new file mode 100644
index 00000000..a81ee4fc
Binary files /dev/null and b/samples/sample6/doc-scans/sample_roll_02.jpg differ
diff --git a/samples/sample6/doc-scans/sample_roll_03.jpg b/samples/sample6/doc-scans/sample_roll_03.jpg
new file mode 100644
index 00000000..94296f4e
Binary files /dev/null and b/samples/sample6/doc-scans/sample_roll_03.jpg differ
diff --git a/samples/sample6/readme.md b/samples/sample6/readme.md
new file mode 100644
index 00000000..2a02ee90
--- /dev/null
+++ b/samples/sample6/readme.md
@@ -0,0 +1,21 @@
+# Demo for feature-based alignment
+
+## Background
+OMR is used to match student roll on final exam scripts. Scripts are scanned using a document scanner and the cover pages are extracted for OMR. Even though a document scanner does not produce any warpped perspective, the alignment is not perfect, causing some rotation and translation in the scans.
+
+The scripts in this sample were specifically selected incorrectly marked scripts to demonstrate how feature-based alignment can correct transformation errors using a reference image. In the actual batch. 156 out of 532 scripts were incorrectly marked. With feature-based alignment, all scripts were correctly marked.
+
+## Usage
+Two template files are given in the sample folder, one with feature-based alignment (template_fb_align), the other without (template_no_fb_align).
+
+## Additional Notes
+
+### Reference Image
+When using a reference image for feature-based alignment, it is better not to have many repeated patterns as it is causes ambiguity when trying to match similar feature points. The bubbles in an OMR form are identical and should not be used for feature-extraction.
+
+Thus, the reference image should be cleared of any bubbles. Forms with lots of text as in this example would be effective.
+
+Note the reference image in this sample was generated from a vector pdf, and not from a scanned blank, producing in a perfectly aligned reference.
+
+### Level adjustment
+The bubbles on the scripts were not shaded dark enough. Thus, a level adjustment was done to bring the black point to 70% to darken the light shading. White point was brought down to 80% to remove the light-grey background in the columns.
diff --git a/samples/sample6/reference.png b/samples/sample6/reference.png
new file mode 100644
index 00000000..5351267f
Binary files /dev/null and b/samples/sample6/reference.png differ
diff --git a/samples/sample6/template.json b/samples/sample6/template.json
new file mode 100644
index 00000000..57605a6c
--- /dev/null
+++ b/samples/sample6/template.json
@@ -0,0 +1,110 @@
+{
+ "pageDimensions": [
+ 2480,
+ 3508
+ ],
+ "bubbleDimensions": [
+ 42,
+ 42
+ ],
+ "preProcessors": [
+ {
+ "name": "Levels",
+ "options": {
+ "low": 0.7,
+ "high": 0.8
+ }
+ },
+ {
+ "name": "GaussianBlur",
+ "options": {
+ "kSize": [
+ 3,
+ 3
+ ],
+ "sigmaX": 0
+ }
+ }
+ ],
+ "customLabels": {
+ "Roll": [
+ "stu",
+ "roll1..7",
+ "check_1",
+ "check_2"
+ ]
+ },
+ "fieldBlocks": {
+ "Check1": {
+ "origin": [
+ 2033,
+ 1290
+ ],
+ "bubblesGap": 50,
+ "labelsGap": 50,
+ "fieldLabels": [
+ "check_1"
+ ],
+ "bubbleValues": [
+ "A",
+ "B",
+ "E",
+ "H",
+ "J",
+ "L",
+ "M"
+ ],
+ "direction": "vertical"
+ },
+ "Check2": {
+ "origin": [
+ 2083,
+ 1290
+ ],
+ "bubblesGap": 50,
+ "labelsGap": 50,
+ "fieldLabels": [
+ "check_2"
+ ],
+ "bubbleValues": [
+ "N",
+ "R",
+ "U",
+ "W",
+ "X",
+ "Y"
+ ],
+ "direction": "vertical"
+ },
+ "Stu": {
+ "origin": [
+ 1636,
+ 1290
+ ],
+ "bubblesGap": 50,
+ "labelsGap": 50,
+ "fieldLabels": [
+ "stu"
+ ],
+ "bubbleValues": [
+ "U",
+ "A",
+ "HT",
+ "GT"
+ ],
+ "direction": "vertical"
+ },
+ "Roll": {
+ "fieldType": "QTYPE_INT",
+ "origin": [
+ 1685,
+ 1290
+ ],
+ "bubblesGap": 50.5,
+ "labelsGap": 50.5,
+ "fieldLabels": [
+ "roll1..7"
+ ]
+ }
+ }
+}
diff --git a/samples/sample6/template_fb_align.json b/samples/sample6/template_fb_align.json
new file mode 100644
index 00000000..7c0c2397
--- /dev/null
+++ b/samples/sample6/template_fb_align.json
@@ -0,0 +1,118 @@
+{
+ "pageDimensions": [
+ 2480,
+ 3508
+ ],
+ "bubbleDimensions": [
+ 42,
+ 42
+ ],
+ "preProcessors": [
+ {
+ "name": "Levels",
+ "options": {
+ "low": 0.7,
+ "high": 0.8
+ }
+ },
+ {
+ "name": "FeatureBasedAlignment",
+ "options": {
+ "reference": "reference.png",
+ "maxFeatures": 1000,
+ "2d": true
+ }
+ },
+ {
+ "name": "GaussianBlur",
+ "options": {
+ "kSize": [
+ 3,
+ 3
+ ],
+ "sigmaX": 0
+ }
+ }
+ ],
+ "customLabels": {
+ "Roll": [
+ "stu",
+ "roll1..7",
+ "check_1",
+ "check_2"
+ ]
+ },
+ "fieldBlocks": {
+ "Check1": {
+ "origin": [
+ 2033,
+ 1290
+ ],
+ "bubblesGap": 50,
+ "labelsGap": 50,
+ "fieldLabels": [
+ "check_1"
+ ],
+ "bubbleValues": [
+ "A",
+ "B",
+ "E",
+ "H",
+ "J",
+ "L",
+ "M"
+ ],
+ "direction": "vertical"
+ },
+ "Check2": {
+ "origin": [
+ 2083,
+ 1290
+ ],
+ "bubblesGap": 50,
+ "labelsGap": 50,
+ "fieldLabels": [
+ "check_2"
+ ],
+ "bubbleValues": [
+ "N",
+ "R",
+ "U",
+ "W",
+ "X",
+ "Y"
+ ],
+ "direction": "vertical"
+ },
+ "Stu": {
+ "origin": [
+ 1636,
+ 1290
+ ],
+ "bubblesGap": 50,
+ "labelsGap": 50,
+ "fieldLabels": [
+ "stu"
+ ],
+ "bubbleValues": [
+ "U",
+ "A",
+ "HT",
+ "GT"
+ ],
+ "direction": "vertical"
+ },
+ "Roll": {
+ "fieldType": "QTYPE_INT",
+ "origin": [
+ 1685,
+ 1290
+ ],
+ "bubblesGap": 50.5,
+ "labelsGap": 50.5,
+ "fieldLabels": [
+ "roll1..7"
+ ]
+ }
+ }
+}
diff --git a/samples/sample6/template_no_fb_align.json b/samples/sample6/template_no_fb_align.json
new file mode 100644
index 00000000..57605a6c
--- /dev/null
+++ b/samples/sample6/template_no_fb_align.json
@@ -0,0 +1,110 @@
+{
+ "pageDimensions": [
+ 2480,
+ 3508
+ ],
+ "bubbleDimensions": [
+ 42,
+ 42
+ ],
+ "preProcessors": [
+ {
+ "name": "Levels",
+ "options": {
+ "low": 0.7,
+ "high": 0.8
+ }
+ },
+ {
+ "name": "GaussianBlur",
+ "options": {
+ "kSize": [
+ 3,
+ 3
+ ],
+ "sigmaX": 0
+ }
+ }
+ ],
+ "customLabels": {
+ "Roll": [
+ "stu",
+ "roll1..7",
+ "check_1",
+ "check_2"
+ ]
+ },
+ "fieldBlocks": {
+ "Check1": {
+ "origin": [
+ 2033,
+ 1290
+ ],
+ "bubblesGap": 50,
+ "labelsGap": 50,
+ "fieldLabels": [
+ "check_1"
+ ],
+ "bubbleValues": [
+ "A",
+ "B",
+ "E",
+ "H",
+ "J",
+ "L",
+ "M"
+ ],
+ "direction": "vertical"
+ },
+ "Check2": {
+ "origin": [
+ 2083,
+ 1290
+ ],
+ "bubblesGap": 50,
+ "labelsGap": 50,
+ "fieldLabels": [
+ "check_2"
+ ],
+ "bubbleValues": [
+ "N",
+ "R",
+ "U",
+ "W",
+ "X",
+ "Y"
+ ],
+ "direction": "vertical"
+ },
+ "Stu": {
+ "origin": [
+ 1636,
+ 1290
+ ],
+ "bubblesGap": 50,
+ "labelsGap": 50,
+ "fieldLabels": [
+ "stu"
+ ],
+ "bubbleValues": [
+ "U",
+ "A",
+ "HT",
+ "GT"
+ ],
+ "direction": "vertical"
+ },
+ "Roll": {
+ "fieldType": "QTYPE_INT",
+ "origin": [
+ 1685,
+ 1290
+ ],
+ "bubblesGap": 50.5,
+ "labelsGap": 50.5,
+ "fieldLabels": [
+ "roll1..7"
+ ]
+ }
+ }
+}
diff --git a/src/__init__.py b/src/__init__.py
new file mode 100644
index 00000000..7fc528b4
--- /dev/null
+++ b/src/__init__.py
@@ -0,0 +1,5 @@
+# https://docs.python.org/3/tutorial/modules.html#:~:text=The%20__init__.py,on%20the%20module%20search%20path.
+from src.logger import logger
+
+# It takes a few seconds for the imports
+logger.info(f"Loading OMRChecker modules...")
diff --git a/src/constants.py b/src/constants.py
new file mode 100644
index 00000000..f1cdc87b
--- /dev/null
+++ b/src/constants.py
@@ -0,0 +1,54 @@
+"""
+
+ OMRChecker
+
+ Author: Udayraj Deshmukh
+ Github: https://github.com/Udayraj123
+
+"""
+from dotmap import DotMap
+
+# Filenames
+TEMPLATE_FILENAME = "template.json"
+EVALUATION_FILENAME = "evaluation.json"
+CONFIG_FILENAME = "config.json"
+
+FIELD_LABEL_NUMBER_REGEX = r"([^\d]+)(\d*)"
+#
+ERROR_CODES = DotMap(
+ {
+ "MULTI_BUBBLE_WARN": 1,
+ "NO_MARKER_ERR": 2,
+ },
+ _dynamic=False,
+)
+
+FIELD_TYPES = {
+ "QTYPE_INT": {
+ "bubbleValues": ["0", "1", "2", "3", "4", "5", "6", "7", "8", "9"],
+ "direction": "vertical",
+ },
+ "QTYPE_INT_FROM_1": {
+ "bubbleValues": ["1", "2", "3", "4", "5", "6", "7", "8", "9", "0"],
+ "direction": "vertical",
+ },
+ "QTYPE_MCQ4": {"bubbleValues": ["A", "B", "C", "D"], "direction": "horizontal"},
+ "QTYPE_MCQ5": {
+ "bubbleValues": ["A", "B", "C", "D", "E"],
+ "direction": "horizontal",
+ },
+ #
+ # You can create and append custom field types here-
+ #
+}
+
+# TODO: move to interaction.py
+TEXT_SIZE = 0.95
+CLR_BLACK = (50, 150, 150)
+CLR_WHITE = (250, 250, 250)
+CLR_GRAY = (130, 130, 130)
+CLR_DARK_GRAY = (100, 100, 100)
+
+# TODO: move to config.json
+GLOBAL_PAGE_THRESHOLD_WHITE = 200
+GLOBAL_PAGE_THRESHOLD_BLACK = 100
diff --git a/src/core.py b/src/core.py
new file mode 100644
index 00000000..d475105f
--- /dev/null
+++ b/src/core.py
@@ -0,0 +1,721 @@
+import os
+from collections import defaultdict
+from typing import Any
+
+import cv2
+import matplotlib.pyplot as plt
+import numpy as np
+
+import src.constants as constants
+from src.logger import logger
+from src.utils.image import CLAHE_HELPER, ImageUtils
+from src.utils.interaction import InteractionUtils
+
+
+class ImageInstanceOps:
+ """Class to hold fine-tuned utilities for a group of images. One instance for each processing directory."""
+
+ save_img_list: Any = defaultdict(list)
+
+ def __init__(self, tuning_config):
+ super().__init__()
+ self.tuning_config = tuning_config
+ self.save_image_level = tuning_config.outputs.save_image_level
+
+ def apply_preprocessors(self, file_path, in_omr, template):
+ tuning_config = self.tuning_config
+ # resize to conform to template
+ in_omr = ImageUtils.resize_util(
+ in_omr,
+ tuning_config.dimensions.processing_width,
+ tuning_config.dimensions.processing_height,
+ )
+
+ # run pre_processors in sequence
+ for pre_processor in template.pre_processors:
+ in_omr = pre_processor.apply_filter(in_omr, file_path)
+ return in_omr
+
+ def read_omr_response(self, template, image, name, save_dir=None):
+ config = self.tuning_config
+ auto_align = config.alignment_params.auto_align
+ try:
+ img = image.copy()
+ # origDim = img.shape[:2]
+ img = ImageUtils.resize_util(
+ img, template.page_dimensions[0], template.page_dimensions[1]
+ )
+ if img.max() > img.min():
+ img = ImageUtils.normalize_util(img)
+ # Processing copies
+ transp_layer = img.copy()
+ final_marked = img.copy()
+
+ morph = img.copy()
+ self.append_save_img(3, morph)
+
+ if auto_align:
+ # Note: clahe is good for morphology, bad for thresholding
+ morph = CLAHE_HELPER.apply(morph)
+ self.append_save_img(3, morph)
+ # Remove shadows further, make columns/boxes darker (less gamma)
+ morph = ImageUtils.adjust_gamma(
+ morph, config.threshold_params.GAMMA_LOW
+ )
+ # TODO: all numbers should come from either constants or config
+ _, morph = cv2.threshold(morph, 220, 220, cv2.THRESH_TRUNC)
+ morph = ImageUtils.normalize_util(morph)
+ self.append_save_img(3, morph)
+ if config.outputs.show_image_level >= 4:
+ InteractionUtils.show("morph1", morph, 0, 1, config)
+
+ # Move them to data class if needed
+ # Overlay Transparencies
+ alpha = 0.65
+ omr_response = {}
+ multi_marked, multi_roll = 0, 0
+
+ # TODO Make this part useful for visualizing status checks
+ # blackVals=[0]
+ # whiteVals=[255]
+
+ if config.outputs.show_image_level >= 5:
+ all_c_box_vals = {"int": [], "mcq": []}
+ # TODO: simplify this logic
+ q_nums = {"int": [], "mcq": []}
+
+ # Find Shifts for the field_blocks --> Before calculating threshold!
+ if auto_align:
+ # print("Begin Alignment")
+ # Open : erode then dilate
+ v_kernel = cv2.getStructuringElement(cv2.MORPH_RECT, (2, 10))
+ morph_v = cv2.morphologyEx(
+ morph, cv2.MORPH_OPEN, v_kernel, iterations=3
+ )
+ _, morph_v = cv2.threshold(morph_v, 200, 200, cv2.THRESH_TRUNC)
+ morph_v = 255 - ImageUtils.normalize_util(morph_v)
+
+ if config.outputs.show_image_level >= 3:
+ InteractionUtils.show(
+ "morphed_vertical", morph_v, 0, 1, config=config
+ )
+
+ # InteractionUtils.show("morph1",morph,0,1,config=config)
+ # InteractionUtils.show("morphed_vertical",morph_v,0,1,config=config)
+
+ self.append_save_img(3, morph_v)
+
+ morph_thr = 60 # for Mobile images, 40 for scanned Images
+ _, morph_v = cv2.threshold(morph_v, morph_thr, 255, cv2.THRESH_BINARY)
+ # kernel best tuned to 5x5 now
+ morph_v = cv2.erode(morph_v, np.ones((5, 5), np.uint8), iterations=2)
+
+ self.append_save_img(3, morph_v)
+ # h_kernel = cv2.getStructuringElement(cv2.MORPH_RECT, (10, 2))
+ # morph_h = cv2.morphologyEx(morph, cv2.MORPH_OPEN, h_kernel, iterations=3)
+ # ret, morph_h = cv2.threshold(morph_h,200,200,cv2.THRESH_TRUNC)
+ # morph_h = 255 - normalize_util(morph_h)
+ # InteractionUtils.show("morph_h",morph_h,0,1,config=config)
+ # _, morph_h = cv2.threshold(morph_h,morph_thr,255,cv2.THRESH_BINARY)
+ # morph_h = cv2.erode(morph_h, np.ones((5,5),np.uint8), iterations = 2)
+ if config.outputs.show_image_level >= 3:
+ InteractionUtils.show(
+ "morph_thr_eroded", morph_v, 0, 1, config=config
+ )
+
+ self.append_save_img(6, morph_v)
+
+ # template relative alignment code
+ for field_block in template.field_blocks:
+ s, d = field_block.origin, field_block.dimensions
+
+ match_col, max_steps, align_stride, thk = map(
+ config.alignment_params.get,
+ [
+ "match_col",
+ "max_steps",
+ "stride",
+ "thickness",
+ ],
+ )
+ shift, steps = 0, 0
+ while steps < max_steps:
+ left_mean = np.mean(
+ morph_v[
+ s[1] : s[1] + d[1],
+ s[0] + shift - thk : -thk + s[0] + shift + match_col,
+ ]
+ )
+ right_mean = np.mean(
+ morph_v[
+ s[1] : s[1] + d[1],
+ s[0]
+ + shift
+ - match_col
+ + d[0]
+ + thk : thk
+ + s[0]
+ + shift
+ + d[0],
+ ]
+ )
+
+ # For demonstration purposes-
+ # if(field_block.name == "int1"):
+ # ret = morph_v.copy()
+ # cv2.rectangle(ret,
+ # (s[0]+shift-thk,s[1]),
+ # (s[0]+shift+thk+d[0],s[1]+d[1]),
+ # constants.CLR_WHITE,
+ # 3)
+ # appendSaveImg(6,ret)
+ # print(shift, left_mean, right_mean)
+ left_shift, right_shift = left_mean > 100, right_mean > 100
+ if left_shift:
+ if right_shift:
+ break
+ else:
+ shift -= align_stride
+ else:
+ if right_shift:
+ shift += align_stride
+ else:
+ break
+ steps += 1
+
+ field_block.shift = shift
+ # print("Aligned field_block: ",field_block.name,"Corrected Shift:",
+ # field_block.shift,", dimensions:", field_block.dimensions,
+ # "origin:", field_block.origin,'\n')
+ # print("End Alignment")
+
+ final_align = None
+ if config.outputs.show_image_level >= 2:
+ initial_align = self.draw_template_layout(img, template, shifted=False)
+ final_align = self.draw_template_layout(
+ img, template, shifted=True, draw_qvals=True
+ )
+ # appendSaveImg(4,mean_vals)
+ self.append_save_img(2, initial_align)
+ self.append_save_img(2, final_align)
+
+ if auto_align:
+ final_align = np.hstack((initial_align, final_align))
+ self.append_save_img(5, img)
+
+ # Get mean bubbleValues n other stats
+ all_q_vals, all_q_strip_arrs, all_q_std_vals = [], [], []
+ total_q_strip_no = 0
+ for field_block in template.field_blocks:
+ box_w, box_h = field_block.bubble_dimensions
+ q_std_vals = []
+ for field_block_bubbles in field_block.traverse_bubbles:
+ q_strip_vals = []
+ for pt in field_block_bubbles:
+ # shifted
+ x, y = (pt.x + field_block.shift, pt.y)
+ rect = [y, y + box_h, x, x + box_w]
+ q_strip_vals.append(
+ cv2.mean(img[rect[0] : rect[1], rect[2] : rect[3]])[0]
+ # detectCross(img, rect) ? 100 : 0
+ )
+ q_std_vals.append(round(np.std(q_strip_vals), 2))
+ all_q_strip_arrs.append(q_strip_vals)
+ # _, _, _ = get_global_threshold(q_strip_vals, "QStrip Plot",
+ # plot_show=False, sort_in_plot=True)
+ # hist = getPlotImg()
+ # InteractionUtils.show("QStrip "+field_block_bubbles[0].field_label, hist, 0, 1,config=config)
+ all_q_vals.extend(q_strip_vals)
+ # print(total_q_strip_no, field_block_bubbles[0].field_label, q_std_vals[len(q_std_vals)-1])
+ total_q_strip_no += 1
+ all_q_std_vals.extend(q_std_vals)
+
+ global_std_thresh, _, _ = self.get_global_threshold(
+ all_q_std_vals
+ ) # , "Q-wise Std-dev Plot", plot_show=True, sort_in_plot=True)
+ # plt.show()
+ # hist = getPlotImg()
+ # InteractionUtils.show("StdHist", hist, 0, 1,config=config)
+
+ # Note: Plotting takes Significant times here --> Change Plotting args
+ # to support show_image_level
+ # , "Mean Intensity Histogram",plot_show=True, sort_in_plot=True)
+ global_thr, _, _ = self.get_global_threshold(all_q_vals, looseness=4)
+
+ logger.info(
+ f"Thresholding: \tglobal_thr: {round(global_thr, 2)} \tglobal_std_THR: {round(global_std_thresh, 2)}\t{'(Looks like a Xeroxed OMR)' if (global_thr == 255) else ''}"
+ )
+ # plt.show()
+ # hist = getPlotImg()
+ # InteractionUtils.show("StdHist", hist, 0, 1,config=config)
+
+ # if(config.outputs.show_image_level>=1):
+ # hist = getPlotImg()
+ # InteractionUtils.show("Hist", hist, 0, 1,config=config)
+ # appendSaveImg(4,hist)
+ # appendSaveImg(5,hist)
+ # appendSaveImg(2,hist)
+
+ per_omr_threshold_avg, total_q_strip_no, total_q_box_no = 0, 0, 0
+ for field_block in template.field_blocks:
+ block_q_strip_no = 1
+ box_w, box_h = field_block.bubble_dimensions
+ shift = field_block.shift
+ s, d = field_block.origin, field_block.dimensions
+ key = field_block.name[:3]
+ # cv2.rectangle(final_marked,(s[0]+shift,s[1]),(s[0]+shift+d[0],
+ # s[1]+d[1]),CLR_BLACK,3)
+ for field_block_bubbles in field_block.traverse_bubbles:
+ # All Black or All White case
+ no_outliers = all_q_std_vals[total_q_strip_no] < global_std_thresh
+ # print(total_q_strip_no, field_block_bubbles[0].field_label,
+ # all_q_std_vals[total_q_strip_no], "no_outliers:", no_outliers)
+ per_q_strip_threshold = self.get_local_threshold(
+ all_q_strip_arrs[total_q_strip_no],
+ global_thr,
+ no_outliers,
+ f"Mean Intensity Histogram for {key}.{field_block_bubbles[0].field_label}.{block_q_strip_no}",
+ config.outputs.show_image_level >= 6,
+ )
+ # print(field_block_bubbles[0].field_label,key,block_q_strip_no, "THR: ",
+ # round(per_q_strip_threshold,2))
+ per_omr_threshold_avg += per_q_strip_threshold
+
+ # Note: Little debugging visualization - view the particular Qstrip
+ # if(
+ # 0
+ # # or "q17" in (field_block_bubbles[0].field_label)
+ # # or (field_block_bubbles[0].field_label+str(block_q_strip_no))=="q15"
+ # ):
+ # st, end = qStrip
+ # InteractionUtils.show("QStrip: "+key+"-"+str(block_q_strip_no),
+ # img[st[1] : end[1], st[0]+shift : end[0]+shift],0,config=config)
+
+ # TODO: get rid of total_q_box_no
+ detected_bubbles = []
+ for bubble in field_block_bubbles:
+ bubble_is_marked = (
+ per_q_strip_threshold > all_q_vals[total_q_box_no]
+ )
+ total_q_box_no += 1
+ if bubble_is_marked:
+ detected_bubbles.append(bubble)
+ x, y, field_value = (
+ bubble.x + field_block.shift,
+ bubble.y,
+ bubble.field_value,
+ )
+ cv2.rectangle(
+ final_marked,
+ (int(x + box_w / 12), int(y + box_h / 12)),
+ (
+ int(x + box_w - box_w / 12),
+ int(y + box_h - box_h / 12),
+ ),
+ constants.CLR_DARK_GRAY,
+ 3,
+ )
+
+ cv2.putText(
+ final_marked,
+ str(field_value),
+ (x, y),
+ cv2.FONT_HERSHEY_SIMPLEX,
+ constants.TEXT_SIZE,
+ (20, 20, 10),
+ int(1 + 3.5 * constants.TEXT_SIZE),
+ )
+ else:
+ cv2.rectangle(
+ final_marked,
+ (int(x + box_w / 10), int(y + box_h / 10)),
+ (
+ int(x + box_w - box_w / 10),
+ int(y + box_h - box_h / 10),
+ ),
+ constants.CLR_GRAY,
+ -1,
+ )
+
+ for bubble in detected_bubbles:
+ field_label, field_value = (
+ bubble.field_label,
+ bubble.field_value,
+ )
+ # Only send rolls multi-marked in the directory
+ multi_marked_local = field_label in omr_response
+ omr_response[field_label] = (
+ (omr_response[field_label] + field_value)
+ if multi_marked_local
+ else field_value
+ )
+ # TODO: generalize this into identifier
+ # multi_roll = multi_marked_local and "Roll" in str(q)
+ multi_marked = multi_marked or multi_marked_local
+
+ if len(detected_bubbles) == 0:
+ field_label = field_block_bubbles[0].field_label
+ omr_response[field_label] = field_block.empty_val
+
+ if config.outputs.show_image_level >= 5:
+ if key in all_c_box_vals:
+ q_nums[key].append(f"{key[:2]}_c{str(block_q_strip_no)}")
+ all_c_box_vals[key].append(
+ all_q_strip_arrs[total_q_strip_no]
+ )
+
+ block_q_strip_no += 1
+ total_q_strip_no += 1
+ # /for field_block
+
+ per_omr_threshold_avg /= total_q_strip_no
+ per_omr_threshold_avg = round(per_omr_threshold_avg, 2)
+ # Translucent
+ cv2.addWeighted(
+ final_marked, alpha, transp_layer, 1 - alpha, 0, final_marked
+ )
+ # Box types
+ if config.outputs.show_image_level >= 6:
+ # plt.draw()
+ f, axes = plt.subplots(len(all_c_box_vals), sharey=True)
+ f.canvas.manager.set_window_title(name)
+ ctr = 0
+ type_name = {
+ "int": "Integer",
+ "mcq": "MCQ",
+ "med": "MED",
+ "rol": "Roll",
+ }
+ for k, boxvals in all_c_box_vals.items():
+ axes[ctr].title.set_text(type_name[k] + " Type")
+ axes[ctr].boxplot(boxvals)
+ # thrline=axes[ctr].axhline(per_omr_threshold_avg,color='red',ls='--')
+ # thrline.set_label("Average THR")
+ axes[ctr].set_ylabel("Intensity")
+ axes[ctr].set_xticklabels(q_nums[k])
+ # axes[ctr].legend()
+ ctr += 1
+ # imshow will do the waiting
+ plt.tight_layout(pad=0.5)
+ plt.show()
+
+ if config.outputs.show_image_level >= 3 and final_align is not None:
+ final_align = ImageUtils.resize_util_h(
+ final_align, int(config.dimensions.display_height)
+ )
+ # [final_align.shape[1],0])
+ InteractionUtils.show(
+ "Template Alignment Adjustment", final_align, 0, 0, config=config
+ )
+
+ if config.outputs.save_detections and save_dir is not None:
+ if multi_roll:
+ save_dir = save_dir.joinpath("_MULTI_")
+ image_path = str(save_dir.joinpath(name))
+ ImageUtils.save_img(image_path, final_marked)
+
+ self.append_save_img(2, final_marked)
+
+ if save_dir is not None:
+ for i in range(config.outputs.save_image_level):
+ self.save_image_stacks(i + 1, name, save_dir)
+
+ return omr_response, final_marked, multi_marked, multi_roll
+
+ except Exception as e:
+ raise e
+
+ @staticmethod
+ def draw_template_layout(img, template, shifted=True, draw_qvals=False, border=-1):
+ img = ImageUtils.resize_util(
+ img, template.page_dimensions[0], template.page_dimensions[1]
+ )
+ final_align = img.copy()
+ for field_block in template.field_blocks:
+ s, d = field_block.origin, field_block.dimensions
+ box_w, box_h = field_block.bubble_dimensions
+ shift = field_block.shift
+ if shifted:
+ cv2.rectangle(
+ final_align,
+ (s[0] + shift, s[1]),
+ (s[0] + shift + d[0], s[1] + d[1]),
+ constants.CLR_BLACK,
+ 3,
+ )
+ else:
+ cv2.rectangle(
+ final_align,
+ (s[0], s[1]),
+ (s[0] + d[0], s[1] + d[1]),
+ constants.CLR_BLACK,
+ 3,
+ )
+ for field_block_bubbles in field_block.traverse_bubbles:
+ for pt in field_block_bubbles:
+ x, y = (pt.x + field_block.shift, pt.y) if shifted else (pt.x, pt.y)
+ cv2.rectangle(
+ final_align,
+ (int(x + box_w / 10), int(y + box_h / 10)),
+ (int(x + box_w - box_w / 10), int(y + box_h - box_h / 10)),
+ constants.CLR_GRAY,
+ border,
+ )
+ if draw_qvals:
+ rect = [y, y + box_h, x, x + box_w]
+ cv2.putText(
+ final_align,
+ f"{int(cv2.mean(img[rect[0] : rect[1], rect[2] : rect[3]])[0])}",
+ (rect[2] + 2, rect[0] + (box_h * 2) // 3),
+ cv2.FONT_HERSHEY_SIMPLEX,
+ 0.6,
+ constants.CLR_BLACK,
+ 2,
+ )
+ if shifted:
+ text_in_px = cv2.getTextSize(
+ field_block.name, cv2.FONT_HERSHEY_SIMPLEX, constants.TEXT_SIZE, 4
+ )
+ cv2.putText(
+ final_align,
+ field_block.name,
+ (int(s[0] + d[0] - text_in_px[0][0]), int(s[1] - text_in_px[0][1])),
+ cv2.FONT_HERSHEY_SIMPLEX,
+ constants.TEXT_SIZE,
+ constants.CLR_BLACK,
+ 4,
+ )
+ return final_align
+
+ def get_global_threshold(
+ self,
+ q_vals_orig,
+ plot_title=None,
+ plot_show=True,
+ sort_in_plot=True,
+ looseness=1,
+ ):
+ """
+ Note: Cannot assume qStrip has only-gray or only-white bg
+ (in which case there is only one jump).
+ So there will be either 1 or 2 jumps.
+ 1 Jump :
+ ......
+ ||||||
+ |||||| <-- risky THR
+ |||||| <-- safe THR
+ ....||||||
+ ||||||||||
+
+ 2 Jumps :
+ ......
+ |||||| <-- wrong THR
+ ....||||||
+ |||||||||| <-- safe THR
+ ..||||||||||
+ ||||||||||||
+
+ The abstract "First LARGE GAP" is perfect for this.
+ Current code is considering ONLY TOP 2 jumps(>= MIN_GAP) to be big,
+ gives the smaller one
+
+ """
+ config = self.tuning_config
+ PAGE_TYPE_FOR_THRESHOLD, MIN_JUMP, JUMP_DELTA = map(
+ config.threshold_params.get,
+ [
+ "PAGE_TYPE_FOR_THRESHOLD",
+ "MIN_JUMP",
+ "JUMP_DELTA",
+ ],
+ )
+
+ global_default_threshold = (
+ constants.GLOBAL_PAGE_THRESHOLD_WHITE
+ if PAGE_TYPE_FOR_THRESHOLD == "white"
+ else constants.GLOBAL_PAGE_THRESHOLD_BLACK
+ )
+
+ # Sort the Q bubbleValues
+ # TODO: Change var name of q_vals
+ q_vals = sorted(q_vals_orig)
+ # Find the FIRST LARGE GAP and set it as threshold:
+ ls = (looseness + 1) // 2
+ l = len(q_vals) - ls
+ max1, thr1 = MIN_JUMP, global_default_threshold
+ for i in range(ls, l):
+ jump = q_vals[i + ls] - q_vals[i - ls]
+ if jump > max1:
+ max1 = jump
+ thr1 = q_vals[i - ls] + jump / 2
+
+ # NOTE: thr2 is deprecated, thus is JUMP_DELTA
+ # Make use of the fact that the JUMP_DELTA(Vertical gap ofc) between
+ # values at detected jumps would be atleast 20
+ max2, thr2 = MIN_JUMP, global_default_threshold
+ # Requires atleast 1 gray box to be present (Roll field will ensure this)
+ for i in range(ls, l):
+ jump = q_vals[i + ls] - q_vals[i - ls]
+ new_thr = q_vals[i - ls] + jump / 2
+ if jump > max2 and abs(thr1 - new_thr) > JUMP_DELTA:
+ max2 = jump
+ thr2 = new_thr
+ # global_thr = min(thr1,thr2)
+ global_thr, j_low, j_high = thr1, thr1 - max1 // 2, thr1 + max1 // 2
+
+ # # For normal images
+ # thresholdRead = 116
+ # if(thr1 > thr2 and thr2 > thresholdRead):
+ # print("Note: taking safer thr line.")
+ # global_thr, j_low, j_high = thr2, thr2 - max2//2, thr2 + max2//2
+
+ if plot_title:
+ _, ax = plt.subplots()
+ ax.bar(range(len(q_vals_orig)), q_vals if sort_in_plot else q_vals_orig)
+ ax.set_title(plot_title)
+ thrline = ax.axhline(global_thr, color="green", ls="--", linewidth=5)
+ thrline.set_label("Global Threshold")
+ thrline = ax.axhline(thr2, color="red", ls=":", linewidth=3)
+ thrline.set_label("THR2 Line")
+ # thrline=ax.axhline(j_low,color='red',ls='-.', linewidth=3)
+ # thrline=ax.axhline(j_high,color='red',ls='-.', linewidth=3)
+ # thrline.set_label("Boundary Line")
+ # ax.set_ylabel("Mean Intensity")
+ ax.set_ylabel("Values")
+ ax.set_xlabel("Position")
+ ax.legend()
+ if plot_show:
+ plt.title(plot_title)
+ plt.show()
+
+ return global_thr, j_low, j_high
+
+ def get_local_threshold(
+ self, q_vals, global_thr, no_outliers, plot_title=None, plot_show=True
+ ):
+ """
+ TODO: Update this documentation too-
+ //No more - Assumption : Colwise background color is uniformly gray or white,
+ but not alternating. In this case there is atmost one jump.
+
+ 0 Jump :
+ <-- safe THR?
+ .......
+ ...|||||||
+ |||||||||| <-- safe THR?
+ // How to decide given range is above or below gray?
+ -> global q_vals shall absolutely help here. Just run same function
+ on total q_vals instead of colwise _//
+ How to decide it is this case of 0 jumps
+
+ 1 Jump :
+ ......
+ ||||||
+ |||||| <-- risky THR
+ |||||| <-- safe THR
+ ....||||||
+ ||||||||||
+
+ """
+ config = self.tuning_config
+ # Sort the Q bubbleValues
+ q_vals = sorted(q_vals)
+
+ # Small no of pts cases:
+ # base case: 1 or 2 pts
+ if len(q_vals) < 3:
+ thr1 = (
+ global_thr
+ if np.max(q_vals) - np.min(q_vals) < config.threshold_params.MIN_GAP
+ else np.mean(q_vals)
+ )
+ else:
+ # qmin, qmax, qmean, qstd = round(np.min(q_vals),2), round(np.max(q_vals),2),
+ # round(np.mean(q_vals),2), round(np.std(q_vals),2)
+ # GVals = [round(abs(q-qmean),2) for q in q_vals]
+ # gmean, gstd = round(np.mean(GVals),2), round(np.std(GVals),2)
+ # # DISCRETION: Pretty critical factor in reading response
+ # # Doesn't work well for small number of values.
+ # DISCRETION = 2.7 # 2.59 was closest hit, 3.0 is too far
+ # L2MaxGap = round(max([abs(g-gmean) for g in GVals]),2)
+ # if(L2MaxGap > DISCRETION*gstd):
+ # no_outliers = False
+
+ # # ^Stackoverflow method
+ # print(field_label, no_outliers,"qstd",round(np.std(q_vals),2), "gstd", gstd,
+ # "Gaps in gvals",sorted([round(abs(g-gmean),2) for g in GVals],reverse=True),
+ # '\t',round(DISCRETION*gstd,2), L2MaxGap)
+
+ # else:
+ # Find the LARGEST GAP and set it as threshold: //(FIRST LARGE GAP)
+ l = len(q_vals) - 1
+ max1, thr1 = config.threshold_params.MIN_JUMP, 255
+ for i in range(1, l):
+ jump = q_vals[i + 1] - q_vals[i - 1]
+ if jump > max1:
+ max1 = jump
+ thr1 = q_vals[i - 1] + jump / 2
+ # print(field_label,q_vals,max1)
+
+ confident_jump = (
+ config.threshold_params.MIN_JUMP
+ + config.threshold_params.CONFIDENT_SURPLUS
+ )
+ # If not confident, then only take help of global_thr
+ if max1 < confident_jump:
+ if no_outliers:
+ # All Black or All White case
+ thr1 = global_thr
+ else:
+ # TODO: Low confidence parameters here
+ pass
+
+ # if(thr1 == 255):
+ # print("Warning: threshold is unexpectedly 255! (Outlier Delta issue?)",plot_title)
+
+ # Make a common plot function to show local and global thresholds
+ if plot_show and plot_title is not None:
+ _, ax = plt.subplots()
+ ax.bar(range(len(q_vals)), q_vals)
+ thrline = ax.axhline(thr1, color="green", ls=("-."), linewidth=3)
+ thrline.set_label("Local Threshold")
+ thrline = ax.axhline(global_thr, color="red", ls=":", linewidth=5)
+ thrline.set_label("Global Threshold")
+ ax.set_title(plot_title)
+ ax.set_ylabel("Bubble Mean Intensity")
+ ax.set_xlabel("Bubble Number(sorted)")
+ ax.legend()
+ # TODO append QStrip to this plot-
+ # appendSaveImg(6,getPlotImg())
+ if plot_show:
+ plt.show()
+ return thr1
+
+ def append_save_img(self, key, img):
+ if self.save_image_level >= int(key):
+ self.save_img_list[key].append(img.copy())
+
+ def save_image_stacks(self, key, filename, save_dir):
+ config = self.tuning_config
+ if self.save_image_level >= int(key) and self.save_img_list[key] != []:
+ name = os.path.splitext(filename)[0]
+ result = np.hstack(
+ tuple(
+ [
+ ImageUtils.resize_util_h(img, config.dimensions.display_height)
+ for img in self.save_img_list[key]
+ ]
+ )
+ )
+ result = ImageUtils.resize_util(
+ result,
+ min(
+ len(self.save_img_list[key]) * config.dimensions.display_width // 3,
+ int(config.dimensions.display_width * 2.5),
+ ),
+ )
+ ImageUtils.save_img(f"{save_dir}stack/{name}_{str(key)}_stack.jpg", result)
+
+ def reset_all_save_img(self):
+ for i in range(self.save_image_level):
+ self.save_img_list[i + 1] = []
diff --git a/src/defaults/__init__.py b/src/defaults/__init__.py
new file mode 100644
index 00000000..62ed2e6f
--- /dev/null
+++ b/src/defaults/__init__.py
@@ -0,0 +1,5 @@
+# https://docs.python.org/3/tutorial/modules.html#:~:text=The%20__init__.py,on%20the%20module%20search%20path.
+# Use all imports relative to root directory
+# (https://chrisyeh96.github.io/2017/08/08/definitive-guide-python-imports.html)
+from src.defaults.config import * # NOQA
+from src.defaults.template import * # NOQA
diff --git a/src/defaults/config.py b/src/defaults/config.py
new file mode 100644
index 00000000..1c6cad3f
--- /dev/null
+++ b/src/defaults/config.py
@@ -0,0 +1,35 @@
+from dotmap import DotMap
+
+CONFIG_DEFAULTS = DotMap(
+ {
+ "dimensions": {
+ "display_height": 2480,
+ "display_width": 1640,
+ "processing_height": 820,
+ "processing_width": 666,
+ },
+ "threshold_params": {
+ "GAMMA_LOW": 0.7,
+ "MIN_GAP": 30,
+ "MIN_JUMP": 25,
+ "CONFIDENT_SURPLUS": 5,
+ "JUMP_DELTA": 30,
+ "PAGE_TYPE_FOR_THRESHOLD": "white",
+ },
+ "alignment_params": {
+ # Note: 'auto_align' enables automatic template alignment, use if the scans show slight misalignments.
+ "auto_align": False,
+ "match_col": 5,
+ "max_steps": 20,
+ "stride": 1,
+ "thickness": 3,
+ },
+ "outputs": {
+ "show_image_level": 0,
+ "save_image_level": 0,
+ "save_detections": True,
+ "filter_out_multimarked_files": False,
+ },
+ },
+ _dynamic=False,
+)
diff --git a/src/defaults/template.py b/src/defaults/template.py
new file mode 100644
index 00000000..d0a2a831
--- /dev/null
+++ b/src/defaults/template.py
@@ -0,0 +1,6 @@
+TEMPLATE_DEFAULTS = {
+ "preProcessors": [],
+ "emptyValue": "",
+ "customLabels": {},
+ "outputColumns": [],
+}
diff --git a/src/entry.py b/src/entry.py
new file mode 100644
index 00000000..5cfcfd95
--- /dev/null
+++ b/src/entry.py
@@ -0,0 +1,370 @@
+"""
+
+ OMRChecker
+
+ Author: Udayraj Deshmukh
+ Github: https://github.com/Udayraj123
+
+"""
+import os
+from csv import QUOTE_NONNUMERIC
+from pathlib import Path
+from time import time
+
+import cv2
+import pandas as pd
+from rich.table import Table
+
+from src import constants
+from src.defaults import CONFIG_DEFAULTS
+from src.evaluation import EvaluationConfig, evaluate_concatenated_response
+from src.logger import console, logger
+from src.template import Template
+from src.utils.file import Paths, setup_dirs_for_paths, setup_outputs_for_template
+from src.utils.image import ImageUtils
+from src.utils.interaction import InteractionUtils, Stats
+from src.utils.parsing import get_concatenated_response, open_config_with_defaults
+
+# Load processors
+STATS = Stats()
+
+
+def entry_point(input_dir, args):
+ if not os.path.exists(input_dir):
+ raise Exception(f"Given input directory does not exist: '{input_dir}'")
+ curr_dir = input_dir
+ return process_dir(input_dir, curr_dir, args)
+
+
+def print_config_summary(
+ curr_dir,
+ omr_files,
+ template,
+ tuning_config,
+ local_config_path,
+ evaluation_config,
+ args,
+):
+ logger.info("")
+ table = Table(title="Current Configurations", show_header=False, show_lines=False)
+ table.add_column("Key", style="cyan", no_wrap=True)
+ table.add_column("Value", style="magenta")
+ table.add_row("Directory Path", f"{curr_dir}")
+ table.add_row("Count of Images", f"{len(omr_files)}")
+ table.add_row("Set Layout Mode ", "ON" if args["setLayout"] else "OFF")
+ table.add_row(
+ "Markers Detection",
+ "ON" if "CropOnMarkers" in template.pre_processors else "OFF",
+ )
+ table.add_row("Auto Alignment", f"{tuning_config.alignment_params.auto_align}")
+ table.add_row("Detected Template Path", f"{template}")
+ if local_config_path:
+ table.add_row("Detected Local Config", f"{local_config_path}")
+ if evaluation_config:
+ table.add_row("Detected Evaluation Config", f"{evaluation_config}")
+
+ table.add_row(
+ "Detected pre-processors",
+ f"{[pp.__class__.__name__ for pp in template.pre_processors]}",
+ )
+ console.print(table, justify="center")
+
+
+def process_dir(
+ root_dir,
+ curr_dir,
+ args,
+ template=None,
+ tuning_config=CONFIG_DEFAULTS,
+ evaluation_config=None,
+):
+ # Update local tuning_config (in current recursion stack)
+ local_config_path = curr_dir.joinpath(constants.CONFIG_FILENAME)
+ if os.path.exists(local_config_path):
+ tuning_config = open_config_with_defaults(local_config_path)
+
+ # Update local template (in current recursion stack)
+ local_template_path = curr_dir.joinpath(constants.TEMPLATE_FILENAME)
+ local_template_exists = os.path.exists(local_template_path)
+ if local_template_exists:
+ template = Template(
+ local_template_path,
+ tuning_config,
+ )
+ # Look for subdirectories for processing
+ subdirs = [d for d in curr_dir.iterdir() if d.is_dir()]
+
+ output_dir = Path(args["output_dir"], curr_dir.relative_to(root_dir))
+ paths = Paths(output_dir)
+
+ # look for images in current dir to process
+ exts = ("*.[pP][nN][gG]", "*.[jJ][pP][gG]", "*.[jJ][pP][eE][gG]")
+ omr_files = sorted([f for ext in exts for f in curr_dir.glob(ext)])
+
+ # Exclude images (take union over all pre_processors)
+ excluded_files = []
+ if template:
+ for pp in template.pre_processors:
+ excluded_files.extend(Path(p) for p in pp.exclude_files())
+
+ local_evaluation_path = curr_dir.joinpath(constants.EVALUATION_FILENAME)
+ if not args["setLayout"] and os.path.exists(local_evaluation_path):
+ if not local_template_exists:
+ logger.warning(
+ f"Found an evaluation file without a parent template file: {local_evaluation_path}"
+ )
+ evaluation_config = EvaluationConfig(
+ curr_dir,
+ local_evaluation_path,
+ template,
+ tuning_config,
+ )
+
+ excluded_files.extend(
+ Path(exclude_file) for exclude_file in evaluation_config.get_exclude_files()
+ )
+
+ omr_files = [f for f in omr_files if f not in excluded_files]
+
+ if omr_files:
+ if not template:
+ logger.error(
+ f"Found images, but no template in the directory tree \
+ of '{curr_dir}'. \nPlace {constants.TEMPLATE_FILENAME} in the \
+ appropriate directory."
+ )
+ raise Exception(
+ f"No template file found in the directory tree of {curr_dir}"
+ )
+
+ setup_dirs_for_paths(paths)
+ outputs_namespace = setup_outputs_for_template(paths, template)
+
+ print_config_summary(
+ curr_dir,
+ omr_files,
+ template,
+ tuning_config,
+ local_config_path,
+ evaluation_config,
+ args,
+ )
+ if args["setLayout"]:
+ show_template_layouts(omr_files, template, tuning_config)
+ else:
+ process_files(
+ omr_files,
+ template,
+ tuning_config,
+ evaluation_config,
+ outputs_namespace,
+ )
+
+ elif not subdirs:
+ # Each subdirectory should have images or should be non-leaf
+ logger.info(
+ f"No valid images or sub-folders found in {curr_dir}.\
+ Empty directories not allowed."
+ )
+
+ # recursively process sub-folders
+ for d in subdirs:
+ process_dir(
+ root_dir,
+ d,
+ args,
+ template,
+ tuning_config,
+ evaluation_config,
+ )
+
+
+def show_template_layouts(omr_files, template, tuning_config):
+ for file_path in omr_files:
+ file_name = file_path.name
+ file_path = str(file_path)
+ in_omr = cv2.imread(file_path, cv2.IMREAD_GRAYSCALE)
+ in_omr = template.image_instance_ops.apply_preprocessors(
+ file_path, in_omr, template
+ )
+ template_layout = template.image_instance_ops.draw_template_layout(
+ in_omr, template, shifted=False, border=2
+ )
+ InteractionUtils.show(
+ f"Template Layout: {file_name}", template_layout, 1, 1, config=tuning_config
+ )
+
+
+def process_files(
+ omr_files,
+ template,
+ tuning_config,
+ evaluation_config,
+ outputs_namespace,
+):
+ start_time = int(time())
+ files_counter = 0
+ STATS.files_not_moved = 0
+
+ for file_path in omr_files:
+ files_counter += 1
+ file_name = file_path.name
+
+ in_omr = cv2.imread(str(file_path), cv2.IMREAD_GRAYSCALE)
+
+ logger.info("")
+ logger.info(
+ f"({files_counter}) Opening image: \t'{file_path}'\tResolution: {in_omr.shape}"
+ )
+
+ template.image_instance_ops.reset_all_save_img()
+
+ template.image_instance_ops.append_save_img(1, in_omr)
+
+ in_omr = template.image_instance_ops.apply_preprocessors(
+ file_path, in_omr, template
+ )
+
+ if in_omr is None:
+ # Error OMR case
+ new_file_path = outputs_namespace.paths.errors_dir.joinpath(file_name)
+ outputs_namespace.OUTPUT_SET.append(
+ [file_name] + outputs_namespace.empty_resp
+ )
+ if check_and_move(
+ constants.ERROR_CODES.NO_MARKER_ERR, file_path, new_file_path
+ ):
+ err_line = [
+ file_name,
+ file_path,
+ new_file_path,
+ "NA",
+ ] + outputs_namespace.empty_resp
+ pd.DataFrame(err_line, dtype=str).T.to_csv(
+ outputs_namespace.files_obj["Errors"],
+ mode="a",
+ quoting=QUOTE_NONNUMERIC,
+ header=False,
+ index=False,
+ )
+ continue
+
+ # uniquify
+ file_id = str(file_name)
+ save_dir = outputs_namespace.paths.save_marked_dir
+ (
+ response_dict,
+ final_marked,
+ multi_marked,
+ _,
+ ) = template.image_instance_ops.read_omr_response(
+ template, image=in_omr, name=file_id, save_dir=save_dir
+ )
+
+ # TODO: move inner try catch here
+ # concatenate roll nos, set unmarked responses, etc
+ omr_response = get_concatenated_response(response_dict, template)
+
+ if (
+ evaluation_config is None
+ or not evaluation_config.get_should_explain_scoring()
+ ):
+ logger.info(f"Read Response: \n{omr_response}")
+
+ score = 0
+ if evaluation_config is not None:
+ score = evaluate_concatenated_response(
+ omr_response, evaluation_config, file_path, outputs_namespace.paths.evaluation_dir
+ )
+ logger.info(
+ f"(/{files_counter}) Graded with score: {round(score, 2)}\t for file: '{file_id}'"
+ )
+ else:
+ logger.info(f"(/{files_counter}) Processed file: '{file_id}'")
+
+ if tuning_config.outputs.show_image_level >= 2:
+ InteractionUtils.show(
+ f"Final Marked Bubbles : '{file_id}'",
+ ImageUtils.resize_util_h(
+ final_marked, int(tuning_config.dimensions.display_height * 1.3)
+ ),
+ 1,
+ 1,
+ config=tuning_config,
+ )
+
+ resp_array = []
+ for k in template.output_columns:
+ resp_array.append(omr_response[k])
+
+ outputs_namespace.OUTPUT_SET.append([file_name] + resp_array)
+
+ if multi_marked == 0 or not tuning_config.outputs.filter_out_multimarked_files:
+ STATS.files_not_moved += 1
+ new_file_path = save_dir.joinpath(file_id)
+ # Enter into Results sheet-
+ results_line = [file_name, file_path, new_file_path, score] + resp_array
+ # Write/Append to results_line file(opened in append mode)
+ pd.DataFrame(results_line, dtype=str).T.to_csv(
+ outputs_namespace.files_obj["Results"],
+ mode="a",
+ quoting=QUOTE_NONNUMERIC,
+ header=False,
+ index=False,
+ )
+ else:
+ # multi_marked file
+ logger.info(f"[{files_counter}] Found multi-marked file: '{file_id}'")
+ new_file_path = outputs_namespace.paths.multi_marked_dir.joinpath(file_name)
+ if check_and_move(
+ constants.ERROR_CODES.MULTI_BUBBLE_WARN, file_path, new_file_path
+ ):
+ mm_line = [file_name, file_path, new_file_path, "NA"] + resp_array
+ pd.DataFrame(mm_line, dtype=str).T.to_csv(
+ outputs_namespace.files_obj["MultiMarked"],
+ mode="a",
+ quoting=QUOTE_NONNUMERIC,
+ header=False,
+ index=False,
+ )
+ # else:
+ # TODO: Add appropriate record handling here
+ # pass
+
+ print_stats(start_time, files_counter, tuning_config)
+
+
+def check_and_move(error_code, file_path, filepath2):
+ # TODO: fix file movement into error/multimarked/invalid etc again
+ STATS.files_not_moved += 1
+ return True
+
+
+def print_stats(start_time, files_counter, tuning_config):
+ time_checking = max(1, round(time() - start_time, 2))
+ log = logger.info
+ log("")
+ log(f"{'Total file(s) moved': <27}: {STATS.files_moved}")
+ log(f"{'Total file(s) not moved': <27}: {STATS.files_not_moved}")
+ log("--------------------------------")
+ log(
+ f"{'Total file(s) processed': <27}: {files_counter} ({'Sum Tallied!' if files_counter == (STATS.files_moved + STATS.files_not_moved) else 'Not Tallying!'})"
+ )
+
+ if tuning_config.outputs.show_image_level <= 0:
+ log(
+ f"\nFinished Checking {files_counter} file(s) in {round(time_checking, 1)} seconds i.e. ~{round(time_checking / 60, 1)} minute(s)."
+ )
+ log(
+ f"{'OMR Processing Rate': <27}: \t ~ {round(time_checking / files_counter, 2)} seconds/OMR"
+ )
+ log(
+ f"{'OMR Processing Speed': <27}: \t ~ {round((files_counter * 60) / time_checking, 2)} OMRs/minute"
+ )
+ else:
+ log(f"\n{'Total script time': <27}: {time_checking} seconds")
+
+ if tuning_config.outputs.show_image_level <= 1:
+ log(
+ "\nTip: To see some awesome visuals, open config.json and increase 'show_image_level'"
+ )
diff --git a/src/evaluation.py b/src/evaluation.py
new file mode 100644
index 00000000..0b567169
--- /dev/null
+++ b/src/evaluation.py
@@ -0,0 +1,546 @@
+import ast
+import os
+import re
+from copy import deepcopy
+from csv import QUOTE_NONNUMERIC
+
+import cv2
+import pandas as pd
+from rich.table import Table
+
+from src.logger import console, logger
+from src.schemas.constants import (
+ BONUS_SECTION_PREFIX,
+ DEFAULT_SECTION_KEY,
+ MARKING_VERDICT_TYPES,
+)
+from src.utils.parsing import (
+ get_concatenated_response,
+ open_evaluation_with_validation,
+ parse_fields,
+ parse_float_or_fraction,
+)
+
+
+class AnswerMatcher:
+ def __init__(self, answer_item, section_marking_scheme):
+ self.section_marking_scheme = section_marking_scheme
+ self.answer_item = answer_item
+ self.answer_type = self.validate_and_get_answer_type(answer_item)
+ self.set_defaults_from_scheme(section_marking_scheme)
+
+ @staticmethod
+ def is_a_marking_score(answer_element):
+ # Note: strict type checking is already done at schema validation level,
+ # Here we focus on overall struct type
+ return type(answer_element) == str or type(answer_element) == int
+
+ @staticmethod
+ def is_standard_answer(answer_element):
+ return type(answer_element) == str and len(answer_element) >= 1
+
+ def validate_and_get_answer_type(self, answer_item):
+ if self.is_standard_answer(answer_item):
+ return "standard"
+ elif type(answer_item) == list:
+ if (
+ # Array of answer elements: ['A', 'B', 'AB']
+ len(answer_item) >= 2
+ and all(
+ self.is_standard_answer(answers_or_score)
+ for answers_or_score in answer_item
+ )
+ ):
+ return "multiple-correct"
+ elif (
+ # Array of two-tuples: [['A', 1], ['B', 1], ['C', 3], ['AB', 2]]
+ len(answer_item) >= 1
+ and all(
+ type(answer_and_score) == list and len(answer_and_score) == 2
+ for answer_and_score in answer_item
+ )
+ and all(
+ self.is_standard_answer(allowed_answer)
+ and self.is_a_marking_score(answer_score)
+ for allowed_answer, answer_score in answer_item
+ )
+ ):
+ return "multiple-correct-weighted"
+
+ logger.critical(
+ f"Unable to determine answer type for answer item: {answer_item}"
+ )
+ raise Exception("Unable to determine answer type")
+
+ def set_defaults_from_scheme(self, section_marking_scheme):
+ answer_type = self.answer_type
+ self.empty_val = section_marking_scheme.empty_val
+ answer_item = self.answer_item
+ self.marking = deepcopy(section_marking_scheme.marking)
+ # TODO: reuse part of parse_scheme_marking here -
+ if answer_type == "standard":
+ # no local overrides
+ pass
+ elif answer_type == "multiple-correct":
+ # override marking scheme scores for each allowed answer
+ for allowed_answer in answer_item:
+ self.marking[f"correct-{allowed_answer}"] = self.marking["correct"]
+ elif answer_type == "multiple-correct-weighted":
+ # Note: No override using marking scheme as answer scores are provided in answer_item
+ for allowed_answer, answer_score in answer_item:
+ self.marking[f"correct-{allowed_answer}"] = parse_float_or_fraction(
+ answer_score
+ )
+
+ def get_marking_scheme(self):
+ return self.section_marking_scheme
+
+ def get_section_explanation(self):
+ answer_type = self.answer_type
+ if answer_type in ["standard", "multiple-correct"]:
+ return self.section_marking_scheme.section_key
+ elif answer_type == "multiple-correct-weighted":
+ return f"Custom: {self.marking}"
+
+ def get_verdict_marking(self, marked_answer):
+ answer_type = self.answer_type
+ question_verdict = "incorrect"
+ if answer_type == "standard":
+ question_verdict = self.get_standard_verdict(marked_answer)
+ elif answer_type == "multiple-correct":
+ question_verdict = self.get_multiple_correct_verdict(marked_answer)
+ elif answer_type == "multiple-correct-weighted":
+ question_verdict = self.get_multiple_correct_weighted_verdict(marked_answer)
+ return question_verdict, self.marking[question_verdict]
+
+ def get_standard_verdict(self, marked_answer):
+ allowed_answer = self.answer_item
+ if marked_answer == self.empty_val:
+ return "unmarked"
+ elif marked_answer == allowed_answer:
+ return "correct"
+ else:
+ return "incorrect"
+
+ def get_multiple_correct_verdict(self, marked_answer):
+ allowed_answers = self.answer_item
+ if marked_answer == self.empty_val:
+ return "unmarked"
+ elif marked_answer in allowed_answers:
+ return f"correct-{marked_answer}"
+ else:
+ return "incorrect"
+
+ def get_multiple_correct_weighted_verdict(self, marked_answer):
+ allowed_answers = [
+ allowed_answer for allowed_answer, _answer_score in self.answer_item
+ ]
+ if marked_answer == self.empty_val:
+ return "unmarked"
+ elif marked_answer in allowed_answers:
+ return f"correct-{marked_answer}"
+ else:
+ return "incorrect"
+
+ def __str__(self):
+ return f"{self.answer_item}"
+
+
+class SectionMarkingScheme:
+ def __init__(self, section_key, section_scheme, empty_val):
+ # TODO: get local empty_val from qblock
+ self.empty_val = empty_val
+ self.section_key = section_key
+ # DEFAULT marking scheme follows a shorthand
+ if section_key == DEFAULT_SECTION_KEY:
+ self.questions = None
+ self.marking = self.parse_scheme_marking(section_scheme)
+ else:
+ self.questions = parse_fields(section_key, section_scheme["questions"])
+ self.marking = self.parse_scheme_marking(section_scheme["marking"])
+
+ def __str__(self):
+ return self.section_key
+
+ def parse_scheme_marking(self, marking):
+ parsed_marking = {}
+ for verdict_type in MARKING_VERDICT_TYPES:
+ verdict_marking = parse_float_or_fraction(marking[verdict_type])
+ if (
+ verdict_marking > 0
+ and verdict_type == "incorrect"
+ and not self.section_key.startswith(BONUS_SECTION_PREFIX)
+ ):
+ logger.warning(
+ f"Found positive marks({round(verdict_marking, 2)}) for incorrect answer in the schema '{self.section_key}'. For Bonus sections, add a prefix 'BONUS_' to them."
+ )
+ parsed_marking[verdict_type] = verdict_marking
+
+ return parsed_marking
+
+ def match_answer(self, marked_answer, answer_matcher):
+ question_verdict, verdict_marking = answer_matcher.get_verdict_marking(
+ marked_answer
+ )
+
+ return verdict_marking, question_verdict
+
+
+class EvaluationConfig:
+ """Note: this instance will be reused for multiple omr sheets"""
+
+ def __init__(self, curr_dir, evaluation_path, template, tuning_config):
+ self.path = evaluation_path
+ evaluation_json = open_evaluation_with_validation(evaluation_path)
+ options, marking_schemes, source_type = map(
+ evaluation_json.get, ["options", "marking_schemes", "source_type"]
+ )
+ self.should_explain_scoring = options.get("should_explain_scoring", False)
+ self.has_non_default_section = False
+ self.exclude_files = []
+ self.enable_evaluation_table_to_csv = options.get(
+ "enable_evaluation_table_to_csv", False
+ )
+
+ if source_type == "csv":
+ csv_path = curr_dir.joinpath(options["answer_key_csv_path"])
+ if not os.path.exists(csv_path):
+ logger.warning(f"Answer key csv does not exist at: '{csv_path}'.")
+
+ answer_key_image_path = options.get("answer_key_image_path", None)
+ if os.path.exists(csv_path):
+ # TODO: CSV parsing/validation for each row with a (qNo, ) pair
+ answer_key = pd.read_csv(
+ csv_path,
+ header=None,
+ names=["question", "answer"],
+ converters={"question": str, "answer": self.parse_answer_column},
+ )
+
+ self.questions_in_order = answer_key["question"].to_list()
+ answers_in_order = answer_key["answer"].to_list()
+ elif not answer_key_image_path:
+ raise Exception(f"Answer key csv not found at '{csv_path}'")
+ else:
+ image_path = str(curr_dir.joinpath(answer_key_image_path))
+ if not os.path.exists(image_path):
+ raise Exception(f"Answer key image not found at '{image_path}'")
+
+ # self.exclude_files.append(image_path)
+
+ logger.debug(
+ f"Attempting to generate answer key from image: '{image_path}'"
+ )
+ # TODO: use a common function for below changes?
+ in_omr = cv2.imread(image_path, cv2.IMREAD_GRAYSCALE)
+ in_omr = template.image_instance_ops.apply_preprocessors(
+ image_path, in_omr, template
+ )
+ if in_omr is None:
+ raise Exception(
+ f"Could not read answer key from image {image_path}"
+ )
+ (
+ response_dict,
+ _final_marked,
+ _multi_marked,
+ _multi_roll,
+ ) = template.image_instance_ops.read_omr_response(
+ template,
+ image=in_omr,
+ name=image_path,
+ save_dir=None,
+ )
+ omr_response = get_concatenated_response(response_dict, template)
+
+ empty_val = template.global_empty_val
+ empty_answer_regex = (
+ rf"{re.escape(empty_val)}+" if empty_val != "" else r"^$"
+ )
+
+ if "questions_in_order" in options:
+ self.questions_in_order = self.parse_questions_in_order(
+ options["questions_in_order"]
+ )
+ empty_answered_questions = [
+ question
+ for question in self.questions_in_order
+ if re.search(empty_answer_regex, omr_response[question])
+ ]
+ if len(empty_answered_questions) > 0:
+ logger.error(
+ f"Found empty answers for questions: {empty_answered_questions}, empty value used: '{empty_val}'"
+ )
+ raise Exception(
+ f"Found empty answers in file '{image_path}'. Please check your template again in the --setLayout mode."
+ )
+ else:
+ logger.warning(
+ f"questions_in_order not provided, proceeding to use non-empty values as answer key"
+ )
+ self.questions_in_order = sorted(
+ question
+ for (question, answer) in omr_response.items()
+ if not re.search(empty_answer_regex, answer)
+ )
+ answers_in_order = [
+ omr_response[question] for question in self.questions_in_order
+ ]
+ # TODO: save the CSV
+ else:
+ self.questions_in_order = self.parse_questions_in_order(
+ options["questions_in_order"]
+ )
+ answers_in_order = options["answers_in_order"]
+
+ self.validate_questions(answers_in_order)
+
+ self.section_marking_schemes, self.question_to_scheme = {}, {}
+ for section_key, section_scheme in marking_schemes.items():
+ section_marking_scheme = SectionMarkingScheme(
+ section_key, section_scheme, template.global_empty_val
+ )
+ if section_key != DEFAULT_SECTION_KEY:
+ self.section_marking_schemes[section_key] = section_marking_scheme
+ for q in section_marking_scheme.questions:
+ # TODO: check the answer key for custom scheme here?
+ self.question_to_scheme[q] = section_marking_scheme
+ self.has_non_default_section = True
+ else:
+ self.default_marking_scheme = section_marking_scheme
+
+ self.validate_marking_schemes()
+
+ self.question_to_answer_matcher = self.parse_answers_and_map_questions(
+ answers_in_order
+ )
+ self.validate_answers(answers_in_order, tuning_config)
+
+ def __str__(self):
+ return str(self.path)
+
+ # Externally called methods have higher abstraction level.
+ def prepare_and_validate_omr_response(self, omr_response):
+ self.reset_explanation_table()
+
+ omr_response_questions = set(omr_response.keys())
+ all_questions = set(self.questions_in_order)
+ missing_questions = sorted(all_questions.difference(omr_response_questions))
+ if len(missing_questions) > 0:
+ logger.critical(f"Missing OMR response for: {missing_questions}")
+ raise Exception(
+ f"Some questions are missing in the OMR response for the given answer key"
+ )
+
+ prefixed_omr_response_questions = set(
+ [k for k in omr_response.keys() if k.startswith("q")]
+ )
+ missing_prefixed_questions = sorted(
+ prefixed_omr_response_questions.difference(all_questions)
+ )
+ if len(missing_prefixed_questions) > 0:
+ logger.warning(
+ f"No answer given for potential questions in OMR response: {missing_prefixed_questions}"
+ )
+
+ def match_answer_for_question(self, current_score, question, marked_answer):
+ answer_matcher = self.question_to_answer_matcher[question]
+ question_verdict, delta = answer_matcher.get_verdict_marking(marked_answer)
+ self.conditionally_add_explanation(
+ answer_matcher,
+ delta,
+ marked_answer,
+ question_verdict,
+ question,
+ current_score,
+ )
+ return delta
+
+ def conditionally_print_explanation(self):
+ if self.should_explain_scoring:
+ console.print(self.explanation_table, justify="center")
+
+ # Explanation Table to CSV
+ def conditionally_save_explanation_csv(self, file_path, evaluation_output_dir):
+ if self.enable_evaluation_table_to_csv:
+ data = {col.header: col._cells for col in self.explanation_table.columns}
+
+ output_path = os.path.join(
+ evaluation_output_dir,
+ f"{file_path.stem}_evaluation.csv",
+ )
+
+ pd.DataFrame(data, dtype=str).to_csv(
+ output_path,
+ mode="a",
+ quoting=QUOTE_NONNUMERIC,
+ index=False,
+ )
+
+ def get_should_explain_scoring(self):
+ return self.should_explain_scoring
+
+ def get_exclude_files(self):
+ return self.exclude_files
+
+ @staticmethod
+ def parse_answer_column(answer_column):
+ if answer_column[0] == "[":
+ # multiple-correct-weighted or multiple-correct
+ parsed_answer = ast.literal_eval(answer_column)
+ elif "," in answer_column:
+ # multiple-correct
+ parsed_answer = answer_column.split(",")
+ else:
+ # single-correct
+ parsed_answer = answer_column
+ return parsed_answer
+
+ def parse_questions_in_order(self, questions_in_order):
+ return parse_fields("questions_in_order", questions_in_order)
+
+ def validate_answers(self, answers_in_order, tuning_config):
+ answer_matcher_map = self.question_to_answer_matcher
+ if tuning_config.outputs.filter_out_multimarked_files:
+ multi_marked_answer = False
+ for question, answer_item in zip(self.questions_in_order, answers_in_order):
+ answer_type = answer_matcher_map[question].answer_type
+ if answer_type == "standard":
+ if len(answer_item) > 1:
+ multi_marked_answer = True
+ if answer_type == "multiple-correct":
+ for single_answer in answer_item:
+ if len(single_answer) > 1:
+ multi_marked_answer = True
+ break
+ if answer_type == "multiple-correct-weighted":
+ for single_answer, _answer_score in answer_item:
+ if len(single_answer) > 1:
+ multi_marked_answer = True
+
+ if multi_marked_answer:
+ raise Exception(
+ f"Provided answer key contains multiple correct answer(s), but config.filter_out_multimarked_files is True. Scoring will get skipped."
+ )
+
+ def validate_questions(self, answers_in_order):
+ questions_in_order = self.questions_in_order
+ len_questions_in_order, len_answers_in_order = len(questions_in_order), len(
+ answers_in_order
+ )
+ if len_questions_in_order != len_answers_in_order:
+ logger.critical(
+ f"questions_in_order({len_questions_in_order}): {questions_in_order}\nanswers_in_order({len_answers_in_order}): {answers_in_order}"
+ )
+ raise Exception(
+ f"Unequal lengths for questions_in_order and answers_in_order ({len_questions_in_order} != {len_answers_in_order})"
+ )
+
+ def validate_marking_schemes(self):
+ section_marking_schemes = self.section_marking_schemes
+ section_questions = set()
+ for section_key, section_scheme in section_marking_schemes.items():
+ if section_key == DEFAULT_SECTION_KEY:
+ continue
+ current_set = set(section_scheme.questions)
+ if not section_questions.isdisjoint(current_set):
+ raise Exception(
+ f"Section '{section_key}' has overlapping question(s) with other sections"
+ )
+ section_questions = section_questions.union(current_set)
+
+ all_questions = set(self.questions_in_order)
+ missing_questions = sorted(section_questions.difference(all_questions))
+ if len(missing_questions) > 0:
+ logger.critical(f"Missing answer key for: {missing_questions}")
+ raise Exception(
+ f"Some questions are missing in the answer key for the given marking scheme"
+ )
+
+ def parse_answers_and_map_questions(self, answers_in_order):
+ question_to_answer_matcher = {}
+ for question, answer_item in zip(self.questions_in_order, answers_in_order):
+ section_marking_scheme = self.get_marking_scheme_for_question(question)
+ answer_matcher = AnswerMatcher(answer_item, section_marking_scheme)
+ question_to_answer_matcher[question] = answer_matcher
+ if (
+ answer_matcher.answer_type == "multiple-correct-weighted"
+ and section_marking_scheme.section_key != DEFAULT_SECTION_KEY
+ ):
+ logger.warning(
+ f"The custom scheme '{section_marking_scheme}' will not apply to question '{question}' as it will use the given answer weights f{answer_item}"
+ )
+ return question_to_answer_matcher
+
+ # Then unfolding lower abstraction levels
+ def reset_explanation_table(self):
+ self.explanation_table = None
+ self.prepare_explanation_table()
+
+ def prepare_explanation_table(self):
+ # TODO: provide a way to export this as csv/pdf
+ if not self.should_explain_scoring:
+ return
+ table = Table(title="Evaluation Explanation Table", show_lines=True)
+ table.add_column("Question")
+ table.add_column("Marked")
+ table.add_column("Answer(s)")
+ table.add_column("Verdict")
+ table.add_column("Delta")
+ table.add_column("Score")
+ # TODO: Add max and min score in explanation (row-wise and total)
+ if self.has_non_default_section:
+ table.add_column("Section")
+ self.explanation_table = table
+
+ def get_marking_scheme_for_question(self, question):
+ return self.question_to_scheme.get(question, self.default_marking_scheme)
+
+ def conditionally_add_explanation(
+ self,
+ answer_matcher,
+ delta,
+ marked_answer,
+ question_verdict,
+ question,
+ current_score,
+ ):
+ if self.should_explain_scoring:
+ next_score = current_score + delta
+ # Conditionally add cells
+ row = [
+ item
+ for item in [
+ question,
+ marked_answer,
+ str(answer_matcher),
+ str.title(question_verdict),
+ str(round(delta, 2)),
+ str(round(next_score, 2)),
+ (
+ answer_matcher.get_section_explanation()
+ if self.has_non_default_section
+ else None
+ ),
+ ]
+ if item is not None
+ ]
+ self.explanation_table.add_row(*row)
+
+
+def evaluate_concatenated_response(
+ concatenated_response, evaluation_config, file_path, evaluation_output_dir
+):
+ evaluation_config.prepare_and_validate_omr_response(concatenated_response)
+ current_score = 0.0
+ for question in evaluation_config.questions_in_order:
+ marked_answer = concatenated_response[question]
+ delta = evaluation_config.match_answer_for_question(
+ current_score, question, marked_answer
+ )
+ current_score += delta
+
+ evaluation_config.conditionally_print_explanation()
+ evaluation_config.conditionally_save_explanation_csv(file_path, evaluation_output_dir)
+
+ return current_score
diff --git a/src/logger.py b/src/logger.py
new file mode 100644
index 00000000..27f44b44
--- /dev/null
+++ b/src/logger.py
@@ -0,0 +1,68 @@
+import logging
+from typing import Union
+
+from rich.console import Console
+from rich.logging import RichHandler
+
+FORMAT = "%(message)s"
+
+# TODO: set logging level from config.json dynamically
+logging.basicConfig(
+ level=logging.INFO,
+ format="%(message)s",
+ datefmt="[%X]",
+ handlers=[RichHandler(rich_tracebacks=True)],
+)
+
+
+class Logger:
+ def __init__(
+ self,
+ name,
+ level: Union[int, str] = logging.NOTSET,
+ message_format="%(message)s",
+ date_format="[%X]",
+ ):
+ self.log = logging.getLogger(name)
+ self.log.setLevel(level)
+ self.log.__format__ = message_format
+ self.log.__date_format__ = date_format
+
+ def debug(self, *msg: object, sep=" ", end="\n") -> None:
+ return self.logutil("debug", *msg, sep=sep)
+
+ def info(self, *msg: object, sep=" ", end="\n") -> None:
+ return self.logutil("info", *msg, sep=sep)
+
+ def warning(self, *msg: object, sep=" ", end="\n") -> None:
+ return self.logutil("warning", *msg, sep=sep)
+
+ def error(self, *msg: object, sep=" ", end="\n") -> None:
+ return self.logutil("error", *msg, sep=sep)
+
+ def critical(self, *msg: object, sep=" ", end="\n") -> None:
+ return self.logutil("critical", *msg, sep=sep)
+
+ def stringify(func):
+ def inner(self, method_type: str, *msg: object, sep=" "):
+ nmsg = []
+ for v in msg:
+ if not isinstance(v, str):
+ v = str(v)
+ nmsg.append(v)
+ return func(self, method_type, *nmsg, sep=sep)
+
+ return inner
+
+ # set stack level to 3 so that the caller of this function is logged, not this function itself.
+ # stack-frame - self.log.debug - logutil - stringify - log method - caller
+ @stringify
+ def logutil(self, method_type: str, *msg: object, sep=" ") -> None:
+ func = getattr(self.log, method_type, None)
+ if not func:
+ raise AttributeError(f"Logger has no method {method_type}")
+ return func(sep.join(msg), stacklevel=4)
+
+
+logger = Logger(__name__)
+console = Console()
diff --git a/src/processors/CropOnMarkers.py b/src/processors/CropOnMarkers.py
new file mode 100644
index 00000000..4867e038
--- /dev/null
+++ b/src/processors/CropOnMarkers.py
@@ -0,0 +1,233 @@
+import os
+
+import cv2
+import numpy as np
+
+from src.logger import logger
+from src.processors.interfaces.ImagePreprocessor import ImagePreprocessor
+from src.utils.image import ImageUtils
+from src.utils.interaction import InteractionUtils
+
+
+class CropOnMarkers(ImagePreprocessor):
+ def __init__(self, *args, **kwargs):
+ super().__init__(*args, **kwargs)
+ config = self.tuning_config
+ marker_ops = self.options
+ self.threshold_circles = []
+ # img_utils = ImageUtils()
+
+ # options with defaults
+ self.marker_path = os.path.join(
+ self.relative_dir, marker_ops.get("relativePath", "omr_marker.jpg")
+ )
+ self.min_matching_threshold = marker_ops.get("min_matching_threshold", 0.3)
+ self.max_matching_variation = marker_ops.get("max_matching_variation", 0.41)
+ self.marker_rescale_range = tuple(
+ int(r) for r in marker_ops.get("marker_rescale_range", (35, 100))
+ )
+ self.marker_rescale_steps = int(marker_ops.get("marker_rescale_steps", 10))
+ self.apply_erode_subtract = marker_ops.get("apply_erode_subtract", True)
+ self.marker = self.load_marker(marker_ops, config)
+
+ def __str__(self):
+ return self.marker_path
+
+ def exclude_files(self):
+ return [self.marker_path]
+
+ def apply_filter(self, image, file_path):
+ config = self.tuning_config
+ image_instance_ops = self.image_instance_ops
+ image_eroded_sub = ImageUtils.normalize_util(
+ image
+ if self.apply_erode_subtract
+ else (image - cv2.erode(image, kernel=np.ones((5, 5)), iterations=5))
+ )
+ # Quads on warped image
+ quads = {}
+ h1, w1 = image_eroded_sub.shape[:2]
+ midh, midw = h1 // 3, w1 // 2
+ origins = [[0, 0], [midw, 0], [0, midh], [midw, midh]]
+ quads[0] = image_eroded_sub[0:midh, 0:midw]
+ quads[1] = image_eroded_sub[0:midh, midw:w1]
+ quads[2] = image_eroded_sub[midh:h1, 0:midw]
+ quads[3] = image_eroded_sub[midh:h1, midw:w1]
+
+ # Draw Quadlines
+ image_eroded_sub[:, midw : midw + 2] = 255
+ image_eroded_sub[midh : midh + 2, :] = 255
+
+ best_scale, all_max_t = self.getBestMatch(image_eroded_sub)
+ if best_scale is None:
+ if config.outputs.show_image_level >= 1:
+ InteractionUtils.show("Quads", image_eroded_sub, config=config)
+ return None
+
+ optimal_marker = ImageUtils.resize_util_h(
+ self.marker, u_height=int(self.marker.shape[0] * best_scale)
+ )
+ _h, w = optimal_marker.shape[:2]
+ centres = []
+ sum_t, max_t = 0, 0
+ quarter_match_log = "Matching Marker: "
+ for k in range(0, 4):
+ res = cv2.matchTemplate(quads[k], optimal_marker, cv2.TM_CCOEFF_NORMED)
+ max_t = res.max()
+ quarter_match_log += f"Quarter{str(k + 1)}: {str(round(max_t, 3))}\t"
+ if (
+ max_t < self.min_matching_threshold
+ or abs(all_max_t - max_t) >= self.max_matching_variation
+ ):
+ logger.error(
+ file_path,
+ "\nError: No circle found in Quad",
+ k + 1,
+ "\n\t min_matching_threshold",
+ self.min_matching_threshold,
+ "\t max_matching_variation",
+ self.max_matching_variation,
+ "\t max_t",
+ max_t,
+ "\t all_max_t",
+ all_max_t,
+ )
+ if config.outputs.show_image_level >= 1:
+ InteractionUtils.show(
+ f"No markers: {file_path}",
+ image_eroded_sub,
+ 0,
+ config=config,
+ )
+ InteractionUtils.show(
+ f"res_Q{str(k + 1)} ({str(max_t)})",
+ res,
+ 1,
+ config=config,
+ )
+ return None
+
+ pt = np.argwhere(res == max_t)[0]
+ pt = [pt[1], pt[0]]
+ pt[0] += origins[k][0]
+ pt[1] += origins[k][1]
+ # print(">>",pt)
+ image = cv2.rectangle(
+ image, tuple(pt), (pt[0] + w, pt[1] + _h), (150, 150, 150), 2
+ )
+ # display:
+ image_eroded_sub = cv2.rectangle(
+ image_eroded_sub,
+ tuple(pt),
+ (pt[0] + w, pt[1] + _h),
+ (50, 50, 50) if self.apply_erode_subtract else (155, 155, 155),
+ 4,
+ )
+ centres.append([pt[0] + w / 2, pt[1] + _h / 2])
+ sum_t += max_t
+
+ logger.info(quarter_match_log)
+ logger.info(f"Optimal Scale: {best_scale}")
+ # analysis data
+ self.threshold_circles.append(sum_t / 4)
+
+ image = ImageUtils.four_point_transform(image, np.array(centres))
+ # appendSaveImg(1,image_eroded_sub)
+ # appendSaveImg(1,image_norm)
+
+ image_instance_ops.append_save_img(2, image_eroded_sub)
+ # Debugging image -
+ # res = cv2.matchTemplate(image_eroded_sub,optimal_marker,cv2.TM_CCOEFF_NORMED)
+ # res[ : , midw:midw+2] = 255
+ # res[ midh:midh+2, : ] = 255
+ # show("Markers Matching",res)
+ if config.outputs.show_image_level >= 2 and config.outputs.show_image_level < 4:
+ image_eroded_sub = ImageUtils.resize_util_h(
+ image_eroded_sub, image.shape[0]
+ )
+ image_eroded_sub[:, -5:] = 0
+ h_stack = np.hstack((image_eroded_sub, image))
+ InteractionUtils.show(
+ f"Warped: {file_path}",
+ ImageUtils.resize_util(
+ h_stack, int(config.dimensions.display_width * 1.6)
+ ),
+ 0,
+ 0,
+ [0, 0],
+ config=config,
+ )
+ # iterations : Tuned to 2.
+ # image_eroded_sub = image_norm - cv2.erode(image_norm, kernel=np.ones((5,5)),iterations=2)
+ return image
+
+ def load_marker(self, marker_ops, config):
+ if not os.path.exists(self.marker_path):
+ logger.error(
+ "Marker not found at path provided in template:",
+ self.marker_path,
+ )
+ exit(31)
+
+ marker = cv2.imread(self.marker_path, cv2.IMREAD_GRAYSCALE)
+
+ if "sheetToMarkerWidthRatio" in marker_ops:
+ marker = ImageUtils.resize_util(
+ marker,
+ config.dimensions.processing_width
+ / int(marker_ops["sheetToMarkerWidthRatio"]),
+ )
+ marker = cv2.GaussianBlur(marker, (5, 5), 0)
+ marker = cv2.normalize(
+ marker, None, alpha=0, beta=255, norm_type=cv2.NORM_MINMAX
+ )
+
+ if self.apply_erode_subtract:
+ marker -= cv2.erode(marker, kernel=np.ones((5, 5)), iterations=5)
+
+ return marker
+
+ # Resizing the marker within scaleRange at rate of descent_per_step to
+ # find the best match.
+ def getBestMatch(self, image_eroded_sub):
+ config = self.tuning_config
+ descent_per_step = (
+ self.marker_rescale_range[1] - self.marker_rescale_range[0]
+ ) // self.marker_rescale_steps
+ _h, _w = self.marker.shape[:2]
+ res, best_scale = None, None
+ all_max_t = 0
+
+ for r0 in np.arange(
+ self.marker_rescale_range[1],
+ self.marker_rescale_range[0],
+ -1 * descent_per_step,
+ ): # reverse order
+ s = float(r0 * 1 / 100)
+ if s == 0.0:
+ continue
+ rescaled_marker = ImageUtils.resize_util_h(
+ self.marker, u_height=int(_h * s)
+ )
+ # res is the black image with white dots
+ res = cv2.matchTemplate(
+ image_eroded_sub, rescaled_marker, cv2.TM_CCOEFF_NORMED
+ )
+
+ max_t = res.max()
+ if all_max_t < max_t:
+ # print('Scale: '+str(s)+', Circle Match: '+str(round(max_t*100,2))+'%')
+ best_scale, all_max_t = s, max_t
+
+ if all_max_t < self.min_matching_threshold:
+ logger.warning(
+ "\tTemplate matching too low! Consider rechecking preProcessors applied before this."
+ )
+ if config.outputs.show_image_level >= 1:
+ InteractionUtils.show("res", res, 1, 0, config=config)
+
+ if best_scale is None:
+ logger.warning(
+ "No matchings for given scaleRange:", self.marker_rescale_range
+ )
+ return best_scale, all_max_t
diff --git a/src/processors/CropPage.py b/src/processors/CropPage.py
new file mode 100644
index 00000000..25629e41
--- /dev/null
+++ b/src/processors/CropPage.py
@@ -0,0 +1,112 @@
+"""
+https://www.pyimagesearch.com/2015/04/06/zero-parameter-automatic-canny-edge-detection-with-python-and-opencv/
+"""
+import cv2
+import numpy as np
+
+from src.logger import logger
+from src.processors.interfaces.ImagePreprocessor import ImagePreprocessor
+from src.utils.image import ImageUtils
+from src.utils.interaction import InteractionUtils
+
+MIN_PAGE_AREA = 80000
+
+
+def normalize(image):
+ return cv2.normalize(image, 0, 255, norm_type=cv2.NORM_MINMAX)
+
+
+def check_max_cosine(approx):
+ # assumes 4 pts present
+ max_cosine = 0
+ min_cosine = 1.5
+ for i in range(2, 5):
+ cosine = abs(angle(approx[i % 4], approx[i - 2], approx[i - 1]))
+ max_cosine = max(cosine, max_cosine)
+ min_cosine = min(cosine, min_cosine)
+
+ if max_cosine >= 0.35:
+ logger.warning("Quadrilateral is not a rectangle.")
+ return False
+ return True
+
+
+def validate_rect(approx):
+ return len(approx) == 4 and check_max_cosine(approx.reshape(4, 2))
+
+
+def angle(p_1, p_2, p_0):
+ dx1 = float(p_1[0] - p_0[0])
+ dy1 = float(p_1[1] - p_0[1])
+ dx2 = float(p_2[0] - p_0[0])
+ dy2 = float(p_2[1] - p_0[1])
+ return (dx1 * dx2 + dy1 * dy2) / np.sqrt(
+ (dx1 * dx1 + dy1 * dy1) * (dx2 * dx2 + dy2 * dy2) + 1e-10
+ )
+
+
+class CropPage(ImagePreprocessor):
+ def __init__(self, *args, **kwargs):
+ super().__init__(*args, **kwargs)
+ cropping_ops = self.options
+ self.morph_kernel = tuple(
+ int(x) for x in cropping_ops.get("morphKernel", [10, 10])
+ )
+
+ def apply_filter(self, image, file_path):
+ image = normalize(cv2.GaussianBlur(image, (3, 3), 0))
+
+ # Resize should be done with another preprocessor is needed
+ sheet = self.find_page(image, file_path)
+ if len(sheet) == 0:
+ logger.error(
+ f"\tError: Paper boundary not found for: '{file_path}'\nHave you accidentally included CropPage preprocessor?"
+ )
+ return None
+
+ logger.info(f"Found page corners: \t {sheet.tolist()}")
+
+ # Warp layer 1
+ image = ImageUtils.four_point_transform(image, sheet)
+
+ # Return preprocessed image
+ return image
+
+ def find_page(self, image, file_path):
+ config = self.tuning_config
+
+ image = normalize(image)
+
+ _ret, image = cv2.threshold(image, 200, 255, cv2.THRESH_TRUNC)
+ image = normalize(image)
+
+ kernel = cv2.getStructuringElement(cv2.MORPH_RECT, self.morph_kernel)
+
+ # Close the small holes, i.e. Complete the edges on canny image
+ closed = cv2.morphologyEx(image, cv2.MORPH_CLOSE, kernel)
+
+ edge = cv2.Canny(closed, 185, 55)
+
+ if config.outputs.show_image_level >= 5:
+ InteractionUtils.show("edge", edge, config=config)
+
+ # findContours returns outer boundaries in CW and inner ones, ACW.
+ cnts = ImageUtils.grab_contours(
+ cv2.findContours(edge, cv2.RETR_LIST, cv2.CHAIN_APPROX_SIMPLE)
+ )
+ # convexHull to resolve disordered curves due to noise
+ cnts = [cv2.convexHull(c) for c in cnts]
+ cnts = sorted(cnts, key=cv2.contourArea, reverse=True)[:5]
+ sheet = []
+ for c in cnts:
+ if cv2.contourArea(c) < MIN_PAGE_AREA:
+ continue
+ peri = cv2.arcLength(c, True)
+ approx = cv2.approxPolyDP(c, epsilon=0.025 * peri, closed=True)
+ if validate_rect(approx):
+ sheet = np.reshape(approx, (4, -1))
+ cv2.drawContours(image, [approx], -1, (0, 255, 0), 2)
+ cv2.drawContours(edge, [approx], -1, (255, 255, 255), 10)
+ break
+
+ return sheet
diff --git a/src/processors/FeatureBasedAlignment.py b/src/processors/FeatureBasedAlignment.py
new file mode 100644
index 00000000..be872924
--- /dev/null
+++ b/src/processors/FeatureBasedAlignment.py
@@ -0,0 +1,94 @@
+"""
+Image based feature alignment
+Credits: https://www.learnopencv.com/image-alignment-feature-based-using-opencv-c-python/
+"""
+import cv2
+import numpy as np
+
+from src.processors.interfaces.ImagePreprocessor import ImagePreprocessor
+from src.utils.image import ImageUtils
+from src.utils.interaction import InteractionUtils
+
+
+class FeatureBasedAlignment(ImagePreprocessor):
+ def __init__(self, *args, **kwargs):
+ super().__init__(*args, **kwargs)
+ options = self.options
+ config = self.tuning_config
+
+ # process reference image
+ self.ref_path = self.relative_dir.joinpath(options["reference"])
+ ref_img = cv2.imread(str(self.ref_path), cv2.IMREAD_GRAYSCALE)
+ self.ref_img = ImageUtils.resize_util(
+ ref_img,
+ config.dimensions.processing_width,
+ config.dimensions.processing_height,
+ )
+ # get options with defaults
+ self.max_features = int(options.get("maxFeatures", 500))
+ self.good_match_percent = options.get("goodMatchPercent", 0.15)
+ self.transform_2_d = options.get("2d", False)
+ # Extract keypoints and description of source image
+ self.orb = cv2.ORB_create(self.max_features)
+ self.to_keypoints, self.to_descriptors = self.orb.detectAndCompute(
+ self.ref_img, None
+ )
+
+ def __str__(self):
+ return self.ref_path.name
+
+ def exclude_files(self):
+ return [self.ref_path]
+
+ def apply_filter(self, image, _file_path):
+ config = self.tuning_config
+ # Convert images to grayscale
+ # im1Gray = cv2.cvtColor(im1, cv2.COLOR_BGR2GRAY)
+ # im2Gray = cv2.cvtColor(im2, cv2.COLOR_BGR2GRAY)
+
+ image = cv2.normalize(image, 0, 255, norm_type=cv2.NORM_MINMAX)
+
+ # Detect ORB features and compute descriptors.
+ from_keypoints, from_descriptors = self.orb.detectAndCompute(image, None)
+
+ # Match features.
+ matcher = cv2.DescriptorMatcher_create(
+ cv2.DESCRIPTOR_MATCHER_BRUTEFORCE_HAMMING
+ )
+
+ # create BFMatcher object (alternate matcher)
+ # matcher = cv2.BFMatcher(cv2.NORM_HAMMING, crossCheck=True)
+
+ matches = np.array(matcher.match(from_descriptors, self.to_descriptors, None))
+
+ # Sort matches by score
+ matches = sorted(matches, key=lambda x: x.distance, reverse=False)
+
+ # Remove not so good matches
+ num_good_matches = int(len(matches) * self.good_match_percent)
+ matches = matches[:num_good_matches]
+
+ # Draw top matches
+ if config.outputs.show_image_level > 2:
+ im_matches = cv2.drawMatches(
+ image, from_keypoints, self.ref_img, self.to_keypoints, matches, None
+ )
+ InteractionUtils.show("Aligning", im_matches, resize=True, config=config)
+
+ # Extract location of good matches
+ points1 = np.zeros((len(matches), 2), dtype=np.float32)
+ points2 = np.zeros((len(matches), 2), dtype=np.float32)
+
+ for i, match in enumerate(matches):
+ points1[i, :] = from_keypoints[match.queryIdx].pt
+ points2[i, :] = self.to_keypoints[match.trainIdx].pt
+
+ # Find homography
+ height, width = self.ref_img.shape
+ if self.transform_2_d:
+ m, _inliers = cv2.estimateAffine2D(points1, points2)
+ return cv2.warpAffine(image, m, (width, height))
+
+ # Use homography
+ h, _mask = cv2.findHomography(points1, points2, cv2.RANSAC)
+ return cv2.warpPerspective(image, h, (width, height))
diff --git a/src/processors/builtins.py b/src/processors/builtins.py
new file mode 100644
index 00000000..faa6288f
--- /dev/null
+++ b/src/processors/builtins.py
@@ -0,0 +1,54 @@
+import cv2
+import numpy as np
+
+from src.processors.interfaces.ImagePreprocessor import ImagePreprocessor
+
+
+class Levels(ImagePreprocessor):
+ def __init__(self, *args, **kwargs):
+ super().__init__(*args, **kwargs)
+ options = self.options
+
+ def output_level(value, low, high, gamma):
+ if value <= low:
+ return 0
+ if value >= high:
+ return 255
+ inv_gamma = 1.0 / gamma
+ return (((value - low) / (high - low)) ** inv_gamma) * 255
+
+ self.gamma = np.array(
+ [
+ output_level(
+ i,
+ int(255 * options.get("low", 0)),
+ int(255 * options.get("high", 1)),
+ options.get("gamma", 1.0),
+ )
+ for i in np.arange(0, 256)
+ ]
+ ).astype("uint8")
+
+ def apply_filter(self, image, _file_path):
+ return cv2.LUT(image, self.gamma)
+
+
+class MedianBlur(ImagePreprocessor):
+ def __init__(self, *args, **kwargs):
+ super().__init__(*args, **kwargs)
+ options = self.options
+ self.kSize = int(options.get("kSize", 5))
+
+ def apply_filter(self, image, _file_path):
+ return cv2.medianBlur(image, self.kSize)
+
+
+class GaussianBlur(ImagePreprocessor):
+ def __init__(self, *args, **kwargs):
+ super().__init__(*args, **kwargs)
+ options = self.options
+ self.kSize = tuple(int(x) for x in options.get("kSize", (3, 3)))
+ self.sigmaX = int(options.get("sigmaX", 0))
+
+ def apply_filter(self, image, _file_path):
+ return cv2.GaussianBlur(image, self.kSize, self.sigmaX)
diff --git a/src/processors/interfaces/ImagePreprocessor.py b/src/processors/interfaces/ImagePreprocessor.py
new file mode 100644
index 00000000..a5cf09a4
--- /dev/null
+++ b/src/processors/interfaces/ImagePreprocessor.py
@@ -0,0 +1,18 @@
+# Use all imports relative to root directory
+from src.processors.manager import Processor
+
+
+class ImagePreprocessor(Processor):
+ """Base class for an extension that applies some preprocessing to the input image"""
+
+ def __init__(self, *args, **kwargs):
+ super().__init__(*args, **kwargs)
+
+ def apply_filter(self, image, filename):
+ """Apply filter to the image and returns modified image"""
+ raise NotImplementedError
+
+ @staticmethod
+ def exclude_files():
+ """Returns a list of file paths that should be excluded from processing"""
+ return []
diff --git a/src/processors/manager.py b/src/processors/manager.py
new file mode 100644
index 00000000..c41e0713
--- /dev/null
+++ b/src/processors/manager.py
@@ -0,0 +1,80 @@
+"""
+Processor/Extension framework
+Adapated from https://github.com/gdiepen/python_processor_example
+"""
+import inspect
+import pkgutil
+
+from src.logger import logger
+
+
+class Processor:
+ """Base class that each processor must inherit from."""
+
+ def __init__(
+ self,
+ options=None,
+ relative_dir=None,
+ image_instance_ops=None,
+ ):
+ self.options = options
+ self.relative_dir = relative_dir
+ self.image_instance_ops = image_instance_ops
+ self.tuning_config = image_instance_ops.tuning_config
+ self.description = "UNKNOWN"
+
+
+class ProcessorManager:
+ """Upon creation, this class will read the processors package for modules
+ that contain a class definition that is inheriting from the Processor class
+ """
+
+ def __init__(self, processors_dir="src.processors"):
+ """Constructor that initiates the reading of all available processors
+ when an instance of the ProcessorCollection object is created
+ """
+ self.processors_dir = processors_dir
+ self.reload_processors()
+
+ @staticmethod
+ def get_name_filter(processor_name):
+ def filter_function(member):
+ return inspect.isclass(member) and member.__module__ == processor_name
+
+ return filter_function
+
+ def reload_processors(self):
+ """Reset the list of all processors and initiate the walk over the main
+ provided processor package to load all available processors
+ """
+ self.processors = {}
+ self.seen_paths = []
+
+ logger.info(f'Loading processors from "{self.processors_dir}"...')
+ self.walk_package(self.processors_dir)
+
+ def walk_package(self, package):
+ """walk the supplied package to retrieve all processors"""
+ imported_package = __import__(package, fromlist=["blah"])
+ loaded_packages = []
+ for _, processor_name, ispkg in pkgutil.walk_packages(
+ imported_package.__path__, imported_package.__name__ + "."
+ ):
+ if not ispkg and processor_name != __name__:
+ processor_module = __import__(processor_name, fromlist=["blah"])
+ # https://stackoverflow.com/a/46206754/6242649
+ clsmembers = inspect.getmembers(
+ processor_module,
+ ProcessorManager.get_name_filter(processor_name),
+ )
+ for _, c in clsmembers:
+ # Only add classes that are a sub class of Processor, but NOT Processor itself
+ if issubclass(c, Processor) & (c is not Processor):
+ self.processors[c.__name__] = c
+ loaded_packages.append(c.__name__)
+
+ logger.info(f"Loaded processors: {loaded_packages}")
+
+
+# Singleton export
+PROCESSOR_MANAGER = ProcessorManager()
diff --git a/src/schemas/__init__.py b/src/schemas/__init__.py
new file mode 100644
index 00000000..3ccb298d
--- /dev/null
+++ b/src/schemas/__init__.py
@@ -0,0 +1,18 @@
+# https://docs.python.org/3/tutorial/modules.html#:~:text=The%20__init__.py,on%20the%20module%20search%20path.
+from jsonschema import Draft202012Validator
+
+from src.schemas.config_schema import CONFIG_SCHEMA
+from src.schemas.evaluation_schema import EVALUATION_SCHEMA
+from src.schemas.template_schema import TEMPLATE_SCHEMA
+
+SCHEMA_JSONS = {
+ "config": CONFIG_SCHEMA,
+ "evaluation": EVALUATION_SCHEMA,
+ "template": TEMPLATE_SCHEMA,
+}
+
+SCHEMA_VALIDATORS = {
+ "config": Draft202012Validator(CONFIG_SCHEMA),
+ "evaluation": Draft202012Validator(EVALUATION_SCHEMA),
+ "template": Draft202012Validator(TEMPLATE_SCHEMA),
+}
diff --git a/src/schemas/config_schema.py b/src/schemas/config_schema.py
new file mode 100644
index 00000000..6eb815f8
--- /dev/null
+++ b/src/schemas/config_schema.py
@@ -0,0 +1,57 @@
+CONFIG_SCHEMA = {
+ "$schema": "https://json-schema.org/draft/2020-12/schema",
+ "$id": "https://github.com/Udayraj123/OMRChecker/tree/master/src/schemas/config-schema.json",
+ "title": "Config Schema",
+ "description": "OMRChecker config schema for custom tuning",
+ "type": "object",
+ "additionalProperties": False,
+ "properties": {
+ "dimensions": {
+ "type": "object",
+ "additionalProperties": False,
+ "properties": {
+ "display_height": {"type": "integer"},
+ "display_width": {"type": "integer"},
+ "processing_height": {"type": "integer"},
+ "processing_width": {"type": "integer"},
+ },
+ },
+ "threshold_params": {
+ "type": "object",
+ "additionalProperties": False,
+ "properties": {
+ "GAMMA_LOW": {"type": "number", "minimum": 0, "maximum": 1},
+ "MIN_GAP": {"type": "integer", "minimum": 10, "maximum": 100},
+ "MIN_JUMP": {"type": "integer", "minimum": 10, "maximum": 100},
+ "CONFIDENT_SURPLUS": {"type": "integer", "minimum": 0, "maximum": 20},
+ "JUMP_DELTA": {"type": "integer", "minimum": 10, "maximum": 100},
+ "PAGE_TYPE_FOR_THRESHOLD": {
+ "enum": ["white", "black"],
+ "type": "string",
+ },
+ },
+ },
+ "alignment_params": {
+ "type": "object",
+ "additionalProperties": False,
+ "properties": {
+ "auto_align": {"type": "boolean"},
+ "match_col": {"type": "integer", "minimum": 0, "maximum": 10},
+ "max_steps": {"type": "integer", "minimum": 1, "maximum": 100},
+ "stride": {"type": "integer", "minimum": 1, "maximum": 10},
+ "thickness": {"type": "integer", "minimum": 1, "maximum": 10},
+ },
+ },
+ "outputs": {
+ "type": "object",
+ "additionalProperties": False,
+ "properties": {
+ "show_image_level": {"type": "integer", "minimum": 0, "maximum": 6},
+ "save_image_level": {"type": "integer", "minimum": 0, "maximum": 6},
+ "save_detections": {"type": "boolean"},
+ # This option moves multimarked files into a separate folder for manual checking, skipping evaluation
+ "filter_out_multimarked_files": {"type": "boolean"},
+ },
+ },
+ },
+}
diff --git a/src/schemas/constants.py b/src/schemas/constants.py
new file mode 100644
index 00000000..ccaa6f38
--- /dev/null
+++ b/src/schemas/constants.py
@@ -0,0 +1,17 @@
+DEFAULT_SECTION_KEY = "DEFAULT"
+
+BONUS_SECTION_PREFIX = "BONUS"
+
+MARKING_VERDICT_TYPES = ["correct", "incorrect", "unmarked"]
+
+ARRAY_OF_STRINGS = {
+ "type": "array",
+ "items": {"type": "string"},
+}
+
+FIELD_STRING_TYPE = {
+ "type": "string",
+ "pattern": "^([^\\.]+|[^\\.\\d]+\\d+\\.{2,3}\\d+)$",
+}
+
+FIELD_STRING_REGEX_GROUPS = r"([^\.\d]+)(\d+)\.{2,3}(\d+)"
diff --git a/src/schemas/evaluation_schema.py b/src/schemas/evaluation_schema.py
new file mode 100644
index 00000000..5f2237fd
--- /dev/null
+++ b/src/schemas/evaluation_schema.py
@@ -0,0 +1,151 @@
+from src.schemas.constants import (
+ ARRAY_OF_STRINGS,
+ DEFAULT_SECTION_KEY,
+ FIELD_STRING_TYPE,
+)
+
+marking_score_regex = "-?(\\d+)(/(\\d+))?"
+
+marking_score = {
+ "oneOf": [
+ {"type": "string", "pattern": marking_score_regex},
+ {"type": "number"},
+ ]
+}
+
+marking_object_properties = {
+ "additionalProperties": False,
+ "required": ["correct", "incorrect", "unmarked"],
+ "type": "object",
+ "properties": {
+ # TODO: can support streak marking if we allow array of marking_scores here
+ "correct": marking_score,
+ "incorrect": marking_score,
+ "unmarked": marking_score,
+ },
+}
+
+EVALUATION_SCHEMA = {
+ "$schema": "https://json-schema.org/draft/2020-12/schema",
+ "$id": "https://github.com/Udayraj123/OMRChecker/tree/master/src/schemas/evaluation-schema.json",
+ "title": "Evaluation Schema",
+ "description": "OMRChecker evaluation schema i.e. the marking scheme",
+ "type": "object",
+ "additionalProperties": True,
+ "required": ["source_type", "options", "marking_schemes"],
+ "properties": {
+ "additionalProperties": False,
+ "source_type": {"type": "string", "enum": ["csv", "custom"]},
+ "options": {"type": "object"},
+ "marking_schemes": {
+ "type": "object",
+ "required": [DEFAULT_SECTION_KEY],
+ "patternProperties": {
+ f"^{DEFAULT_SECTION_KEY}$": marking_object_properties,
+ f"^(?!{DEFAULT_SECTION_KEY}$).*": {
+ "additionalProperties": False,
+ "required": ["marking", "questions"],
+ "type": "object",
+ "properties": {
+ "questions": {
+ "oneOf": [
+ FIELD_STRING_TYPE,
+ {
+ "type": "array",
+ "items": FIELD_STRING_TYPE,
+ },
+ ]
+ },
+ "marking": marking_object_properties,
+ },
+ },
+ },
+ },
+ },
+ "allOf": [
+ {
+ "if": {"properties": {"source_type": {"const": "csv"}}},
+ "then": {
+ "properties": {
+ "options": {
+ "additionalProperties": False,
+ "required": ["answer_key_csv_path"],
+ "dependentRequired": {
+ "answer_key_image_path": [
+ "answer_key_csv_path",
+ "questions_in_order",
+ ]
+ },
+ "type": "object",
+ "properties": {
+ "should_explain_scoring": {"type": "boolean"},
+ "answer_key_csv_path": {"type": "string"},
+ "answer_key_image_path": {"type": "string"},
+ "questions_in_order": ARRAY_OF_STRINGS,
+ },
+ }
+ }
+ },
+ },
+ {
+ "if": {"properties": {"source_type": {"const": "custom"}}},
+ "then": {
+ "properties": {
+ "options": {
+ "additionalProperties": False,
+ "required": ["answers_in_order", "questions_in_order"],
+ "type": "object",
+ "properties": {
+ "should_explain_scoring": {"type": "boolean"},
+ "answers_in_order": {
+ "oneOf": [
+ {
+ "type": "array",
+ "items": {
+ "oneOf": [
+ # "standard": single correct, multi-marked single-correct
+ # Example: "q1" --> '67'
+ {"type": "string"},
+ # "multiple-correct": multiple-correct (for ambiguous/bonus questions)
+ # Example: "q1" --> [ 'A', 'B' ]
+ {
+ "type": "array",
+ "items": {"type": "string"},
+ "minItems": 2,
+ },
+ # "multiple-correct-weighted": array of answer-wise weights (marking scheme not applicable)
+ # Example 1: "q1" --> [['A', 1], ['B', 2], ['C', 3]] or
+ # Example 2: "q2" --> [['A', 1], ['B', 1], ['AB', 2]]
+ {
+ "type": "array",
+ "items": {
+ "type": "array",
+ "items": False,
+ "minItems": 2,
+ "maxItems": 2,
+ "prefixItems": [
+ {"type": "string"},
+ marking_score,
+ ],
+ },
+ },
+ # Multiple-correct with custom marking scheme
+ # ["A", ["1", "2", "3"]],
+ # [["A", "B", "AB"], ["1", "2", "3"]]
+ ],
+ },
+ },
+ ]
+ },
+ "questions_in_order": ARRAY_OF_STRINGS,
+ "enable_evaluation_table_to_csv": {
+ "type": "boolean",
+ "default": False,
+ },
+ },
+ }
+ }
+ },
+ },
+ ],
+}
diff --git a/src/schemas/template_schema.py b/src/schemas/template_schema.py
new file mode 100644
index 00000000..451d499d
--- /dev/null
+++ b/src/schemas/template_schema.py
@@ -0,0 +1,226 @@
+from src.constants import FIELD_TYPES
+from src.schemas.constants import ARRAY_OF_STRINGS, FIELD_STRING_TYPE
+
+positive_number = {"type": "number", "minimum": 0}
+positive_integer = {"type": "integer", "minimum": 0}
+two_positive_integers = {
+ "type": "array",
+ "prefixItems": [
+ positive_integer,
+ positive_integer,
+ ],
+ "maxItems": 2,
+ "minItems": 2,
+}
+two_positive_numbers = {
+ "type": "array",
+ "prefixItems": [
+ positive_number,
+ positive_number,
+ ],
+ "maxItems": 2,
+ "minItems": 2,
+}
+zero_to_one_number = {
+ "type": "number",
+ "minimum": 0,
+ "maximum": 1,
+}
+
+TEMPLATE_SCHEMA = {
+ "$schema": "https://json-schema.org/draft/2020-12/schema",
+ "$id": "https://github.com/Udayraj123/OMRChecker/tree/master/src/schemas/template-schema.json",
+ "title": "Template Validation Schema",
+ "description": "OMRChecker input template schema",
+ "type": "object",
+ "required": [
+ "bubbleDimensions",
+ "pageDimensions",
+ "preProcessors",
+ "fieldBlocks",
+ ],
+ "additionalProperties": False,
+ "properties": {
+ "bubbleDimensions": {
+ **two_positive_integers,
+ "description": "The dimensions of the overlay bubble area: [width, height]",
+ },
+ "customLabels": {
+ "description": "The customLabels contain fields that need to be joined together before generating the results sheet",
+ "type": "object",
+ "patternProperties": {
+ "^.*$": {"type": "array", "items": FIELD_STRING_TYPE}
+ },
+ },
+ "outputColumns": {
+ "type": "array",
+ "items": FIELD_STRING_TYPE,
+ "description": "The ordered list of columns to be contained in the output csv(default order: alphabetical)",
+ },
+ "pageDimensions": {
+ **two_positive_integers,
+ "description": "The dimensions(width, height) to which the page will be resized to before applying template",
+ },
+ "preProcessors": {
+ "description": "Custom configuration values to use in the template's directory",
+ "type": "array",
+ "items": {
+ "type": "object",
+ "properties": {
+ "name": {
+ "type": "string",
+ "enum": [
+ "CropOnMarkers",
+ "CropPage",
+ "FeatureBasedAlignment",
+ "GaussianBlur",
+ "Levels",
+ "MedianBlur",
+ ],
+ },
+ },
+ "required": ["name", "options"],
+ "allOf": [
+ {
+ "if": {"properties": {"name": {"const": "CropOnMarkers"}}},
+ "then": {
+ "properties": {
+ "options": {
+ "type": "object",
+ "additionalProperties": False,
+ "properties": {
+ "apply_erode_subtract": {"type": "boolean"},
+ "marker_rescale_range": two_positive_numbers,
+ "marker_rescale_steps": {"type": "number"},
+ "max_matching_variation": {"type": "number"},
+ "min_matching_threshold": {"type": "number"},
+ "relativePath": {"type": "string"},
+ "sheetToMarkerWidthRatio": {"type": "number"},
+ },
+ "required": ["relativePath"],
+ }
+ }
+ },
+ },
+ {
+ "if": {
+ "properties": {"name": {"const": "FeatureBasedAlignment"}}
+ },
+ "then": {
+ "properties": {
+ "options": {
+ "type": "object",
+ "additionalProperties": False,
+ "properties": {
+ "2d": {"type": "boolean"},
+ "goodMatchPercent": {"type": "number"},
+ "maxFeatures": {"type": "integer"},
+ "reference": {"type": "string"},
+ },
+ "required": ["reference"],
+ }
+ }
+ },
+ },
+ {
+ "if": {"properties": {"name": {"const": "Levels"}}},
+ "then": {
+ "properties": {
+ "options": {
+ "type": "object",
+ "additionalProperties": False,
+ "properties": {
+ "gamma": zero_to_one_number,
+ "high": zero_to_one_number,
+ "low": zero_to_one_number,
+ },
+ }
+ }
+ },
+ },
+ {
+ "if": {"properties": {"name": {"const": "MedianBlur"}}},
+ "then": {
+ "properties": {
+ "options": {
+ "type": "object",
+ "additionalProperties": False,
+ "properties": {"kSize": {"type": "integer"}},
+ }
+ }
+ },
+ },
+ {
+ "if": {"properties": {"name": {"const": "GaussianBlur"}}},
+ "then": {
+ "properties": {
+ "options": {
+ "type": "object",
+ "additionalProperties": False,
+ "properties": {
+ "kSize": two_positive_integers,
+ "sigmaX": {"type": "number"},
+ },
+ }
+ }
+ },
+ },
+ {
+ "if": {"properties": {"name": {"const": "CropPage"}}},
+ "then": {
+ "properties": {
+ "options": {
+ "type": "object",
+ "additionalProperties": False,
+ "properties": {
+ "morphKernel": two_positive_integers
+ },
+ }
+ }
+ },
+ },
+ ],
+ },
+ },
+ "fieldBlocks": {
+ "description": "The fieldBlocks denote small groups of adjacent fields",
+ "type": "object",
+ "patternProperties": {
+ "^.*$": {
+ "type": "object",
+ "required": [
+ "origin",
+ "bubblesGap",
+ "labelsGap",
+ "fieldLabels",
+ ],
+ "oneOf": [
+ {"required": ["fieldType"]},
+ {"required": ["bubbleValues", "direction"]},
+ ],
+ "properties": {
+ "bubbleDimensions": two_positive_numbers,
+ "bubblesGap": positive_number,
+ "bubbleValues": ARRAY_OF_STRINGS,
+ "direction": {
+ "type": "string",
+ "enum": ["horizontal", "vertical"],
+ },
+ "emptyValue": {"type": "string"},
+ "fieldLabels": {"type": "array", "items": FIELD_STRING_TYPE},
+ "labelsGap": positive_number,
+ "origin": two_positive_integers,
+ "fieldType": {
+ "type": "string",
+ "enum": list(FIELD_TYPES.keys()),
+ },
+ },
+ }
+ },
+ },
+ "emptyValue": {
+ "description": "The value to be used in case of empty bubble detected at global level.",
+ "type": "string",
+ },
+ },
+}
diff --git a/src/template.py b/src/template.py
new file mode 100644
index 00000000..0acc4672
--- /dev/null
+++ b/src/template.py
@@ -0,0 +1,327 @@
+"""
+
+ OMRChecker
+
+ Author: Udayraj Deshmukh
+ Github: https://github.com/Udayraj123
+
+"""
+from src.constants import FIELD_TYPES
+from src.core import ImageInstanceOps
+from src.logger import logger
+from src.processors.manager import PROCESSOR_MANAGER
+from src.utils.parsing import (
+ custom_sort_output_columns,
+ open_template_with_defaults,
+ parse_fields,
+)
+
+
+class Template:
+ def __init__(self, template_path, tuning_config):
+ self.path = template_path
+ self.image_instance_ops = ImageInstanceOps(tuning_config)
+
+ json_object = open_template_with_defaults(template_path)
+ (
+ custom_labels_object,
+ field_blocks_object,
+ output_columns_array,
+ pre_processors_object,
+ self.bubble_dimensions,
+ self.global_empty_val,
+ self.options,
+ self.page_dimensions,
+ ) = map(
+ json_object.get,
+ [
+ "customLabels",
+ "fieldBlocks",
+ "outputColumns",
+ "preProcessors",
+ "bubbleDimensions",
+ "emptyValue",
+ "options",
+ "pageDimensions",
+ ],
+ )
+
+ self.parse_output_columns(output_columns_array)
+ self.setup_pre_processors(pre_processors_object, template_path.parent)
+ self.setup_field_blocks(field_blocks_object)
+ self.parse_custom_labels(custom_labels_object)
+
+ non_custom_columns, all_custom_columns = (
+ list(self.non_custom_labels),
+ list(custom_labels_object.keys()),
+ )
+
+ if len(self.output_columns) == 0:
+ self.fill_output_columns(non_custom_columns, all_custom_columns)
+
+ self.validate_template_columns(non_custom_columns, all_custom_columns)
+
+ def parse_output_columns(self, output_columns_array):
+ self.output_columns = parse_fields(f"Output Columns", output_columns_array)
+
+ def setup_pre_processors(self, pre_processors_object, relative_dir):
+ # load image pre_processors
+ self.pre_processors = []
+ for pre_processor in pre_processors_object:
+ ProcessorClass = PROCESSOR_MANAGER.processors[pre_processor["name"]]
+ pre_processor_instance = ProcessorClass(
+ options=pre_processor["options"],
+ relative_dir=relative_dir,
+ image_instance_ops=self.image_instance_ops,
+ )
+ self.pre_processors.append(pre_processor_instance)
+
+ def setup_field_blocks(self, field_blocks_object):
+ # Add field_blocks
+ self.field_blocks = []
+ self.all_parsed_labels = set()
+ for block_name, field_block_object in field_blocks_object.items():
+ self.parse_and_add_field_block(block_name, field_block_object)
+
+ def parse_custom_labels(self, custom_labels_object):
+ all_parsed_custom_labels = set()
+ self.custom_labels = {}
+ for custom_label, label_strings in custom_labels_object.items():
+ parsed_labels = parse_fields(f"Custom Label: {custom_label}", label_strings)
+ parsed_labels_set = set(parsed_labels)
+ self.custom_labels[custom_label] = parsed_labels
+
+ missing_custom_labels = sorted(
+ parsed_labels_set.difference(self.all_parsed_labels)
+ )
+ if len(missing_custom_labels) > 0:
+ logger.critical(
+ f"For '{custom_label}', Missing labels - {missing_custom_labels}"
+ )
+ raise Exception(
+ f"Missing field block label(s) in the given template for {missing_custom_labels} from '{custom_label}'"
+ )
+
+ if not all_parsed_custom_labels.isdisjoint(parsed_labels_set):
+ # Note: this can be made a warning, but it's a choice
+ logger.critical(
+ f"field strings overlap for labels: {label_strings} and existing custom labels: {all_parsed_custom_labels}"
+ )
+ raise Exception(
+ f"The field strings for custom label '{custom_label}' overlap with other existing custom labels"
+ )
+
+ all_parsed_custom_labels.update(parsed_labels)
+
+ self.non_custom_labels = self.all_parsed_labels.difference(
+ all_parsed_custom_labels
+ )
+
+ def fill_output_columns(self, non_custom_columns, all_custom_columns):
+ all_template_columns = non_custom_columns + all_custom_columns
+ # Typical case: sort alpha-numerical (natural sort)
+ self.output_columns = sorted(
+ all_template_columns, key=custom_sort_output_columns
+ )
+
+ def validate_template_columns(self, non_custom_columns, all_custom_columns):
+ output_columns_set = set(self.output_columns)
+ all_custom_columns_set = set(all_custom_columns)
+
+ missing_output_columns = sorted(
+ output_columns_set.difference(all_custom_columns_set).difference(
+ self.all_parsed_labels
+ )
+ )
+ if len(missing_output_columns) > 0:
+ logger.critical(f"Missing output columns: {missing_output_columns}")
+ raise Exception(
+ f"Some columns are missing in the field blocks for the given output columns"
+ )
+
+ all_template_columns_set = set(non_custom_columns + all_custom_columns)
+ missing_label_columns = sorted(
+ all_template_columns_set.difference(output_columns_set)
+ )
+ if len(missing_label_columns) > 0:
+ logger.warning(
+ f"Some label columns are not covered in the given output columns: {missing_label_columns}"
+ )
+
+ def parse_and_add_field_block(self, block_name, field_block_object):
+ field_block_object = self.pre_fill_field_block(field_block_object)
+ block_instance = FieldBlock(block_name, field_block_object)
+ self.field_blocks.append(block_instance)
+ self.validate_parsed_labels(field_block_object["fieldLabels"], block_instance)
+
+ def pre_fill_field_block(self, field_block_object):
+ if "fieldType" in field_block_object:
+ field_block_object = {
+ **field_block_object,
+ **FIELD_TYPES[field_block_object["fieldType"]],
+ }
+ else:
+ field_block_object = {**field_block_object, "fieldType": "__CUSTOM__"}
+
+ return {
+ "direction": "vertical",
+ "emptyValue": self.global_empty_val,
+ "bubbleDimensions": self.bubble_dimensions,
+ **field_block_object,
+ }
+
+ def validate_parsed_labels(self, field_labels, block_instance):
+ parsed_field_labels, block_name = (
+ block_instance.parsed_field_labels,
+ block_instance.name,
+ )
+ field_labels_set = set(parsed_field_labels)
+ if not self.all_parsed_labels.isdisjoint(field_labels_set):
+ # Note: in case of two fields pointing to same column, use a custom column instead of same field labels.
+ logger.critical(
+ f"An overlap found between field string: {field_labels} in block '{block_name}' and existing labels: {self.all_parsed_labels}"
+ )
+ raise Exception(
+ f"The field strings for field block {block_name} overlap with other existing fields"
+ )
+ self.all_parsed_labels.update(field_labels_set)
+
+ page_width, page_height = self.page_dimensions
+ block_width, block_height = block_instance.dimensions
+ [block_start_x, block_start_y] = block_instance.origin
+
+ block_end_x, block_end_y = (
+ block_start_x + block_width,
+ block_start_y + block_height,
+ )
+
+ if (
+ block_end_x >= page_width
+ or block_end_y >= page_height
+ or block_start_x < 0
+ or block_start_y < 0
+ ):
+ raise Exception(
+ f"Overflowing field block '{block_name}' with origin {block_instance.origin} and dimensions {block_instance.dimensions} in template with dimensions {self.page_dimensions}"
+ )
+
+ def __str__(self):
+ return str(self.path)
+
+
+class FieldBlock:
+ def __init__(self, block_name, field_block_object):
+ self.name = block_name
+ self.shift = 0
+ self.setup_field_block(field_block_object)
+
+ def setup_field_block(self, field_block_object):
+ # case mapping
+ (
+ bubble_dimensions,
+ bubble_values,
+ bubbles_gap,
+ direction,
+ field_labels,
+ field_type,
+ labels_gap,
+ origin,
+ self.empty_val,
+ ) = map(
+ field_block_object.get,
+ [
+ "bubbleDimensions",
+ "bubbleValues",
+ "bubblesGap",
+ "direction",
+ "fieldLabels",
+ "fieldType",
+ "labelsGap",
+ "origin",
+ "emptyValue",
+ ],
+ )
+ self.parsed_field_labels = parse_fields(
+ f"Field Block Labels: {self.name}", field_labels
+ )
+ self.origin = origin
+ self.bubble_dimensions = bubble_dimensions
+ self.calculate_block_dimensions(
+ bubble_dimensions,
+ bubble_values,
+ bubbles_gap,
+ direction,
+ labels_gap,
+ )
+ self.generate_bubble_grid(
+ bubble_values,
+ bubbles_gap,
+ direction,
+ field_type,
+ labels_gap,
+ )
+
+ def calculate_block_dimensions(
+ self,
+ bubble_dimensions,
+ bubble_values,
+ bubbles_gap,
+ direction,
+ labels_gap,
+ ):
+ _h, _v = (1, 0) if (direction == "vertical") else (0, 1)
+
+ values_dimension = int(
+ bubbles_gap * (len(bubble_values) - 1) + bubble_dimensions[_h]
+ )
+ fields_dimension = int(
+ labels_gap * (len(self.parsed_field_labels) - 1) + bubble_dimensions[_v]
+ )
+ self.dimensions = (
+ [fields_dimension, values_dimension]
+ if (direction == "vertical")
+ else [values_dimension, fields_dimension]
+ )
+
+ def generate_bubble_grid(
+ self,
+ bubble_values,
+ bubbles_gap,
+ direction,
+ field_type,
+ labels_gap,
+ ):
+ _h, _v = (1, 0) if (direction == "vertical") else (0, 1)
+ self.traverse_bubbles = []
+ # Generate the bubble grid
+ lead_point = [float(self.origin[0]), float(self.origin[1])]
+ for field_label in self.parsed_field_labels:
+ bubble_point = lead_point.copy()
+ field_bubbles = []
+ for bubble_value in bubble_values:
+ field_bubbles.append(
+ Bubble(bubble_point.copy(), field_label, field_type, bubble_value)
+ )
+ bubble_point[_h] += bubbles_gap
+ self.traverse_bubbles.append(field_bubbles)
+ lead_point[_v] += labels_gap
+
+
+class Bubble:
+ """
+ Container for a Point Box on the OMR
+
+ field_label is the point's property- field to which this point belongs to
+ It can be used as a roll number column as well. (eg roll1)
+ It can also correspond to a single digit of integer type Q (eg q5d1)
+ """
+
+ def __init__(self, pt, field_label, field_type, field_value):
+ self.x = round(pt[0])
+ self.y = round(pt[1])
+ self.field_label = field_label
+ self.field_type = field_type
+ self.field_value = field_value
+
+ def __str__(self):
+ return str([self.x, self.y])
diff --git a/src/tests/__init__.py b/src/tests/__init__.py
new file mode 100644
index 00000000..855c16a5
--- /dev/null
+++ b/src/tests/__init__.py
@@ -0,0 +1 @@
+# https://stackoverflow.com/a/50169991/6242649
diff --git a/src/tests/__snapshots__/test_all_samples.ambr b/src/tests/__snapshots__/test_all_samples.ambr
new file mode 100644
index 00000000..3049cd3c
--- /dev/null
+++ b/src/tests/__snapshots__/test_all_samples.ambr
@@ -0,0 +1,302 @@
+# serializer version: 1
+# name: test_run_answer_key_using_csv
+ dict({
+ 'Manual/ErrorFiles.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5"
+
+ ''',
+ 'Manual/MultiMarkedFiles.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5"
+
+ ''',
+ 'Results/Results_05AM.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5"
+ "adrian_omr.png","samples/answer-key/using-csv/adrian_omr.png","outputs/answer-key/using-csv/CheckedOMRs/adrian_omr.png","5.0","C","E","A","B","B"
+
+ ''',
+ })
+# ---
+# name: test_run_answer_key_weighted_answers
+ dict({
+ 'images/Manual/ErrorFiles.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5"
+
+ ''',
+ 'images/Manual/MultiMarkedFiles.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5"
+
+ ''',
+ 'images/Results/Results_05AM.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5"
+ "adrian_omr.png","samples/answer-key/weighted-answers/images/adrian_omr.png","outputs/answer-key/weighted-answers/images/CheckedOMRs/adrian_omr.png","5.5","B","E","A","C","B"
+ "adrian_omr_2.png","samples/answer-key/weighted-answers/images/adrian_omr_2.png","outputs/answer-key/weighted-answers/images/CheckedOMRs/adrian_omr_2.png","10.0","C","E","A","B","B"
+
+ ''',
+ })
+# ---
+# name: test_run_community_Antibodyy
+ dict({
+ 'Manual/ErrorFiles.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5","q6"
+
+ ''',
+ 'Manual/MultiMarkedFiles.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5","q6"
+
+ ''',
+ 'Results/Results_05AM.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5","q6"
+ "simple_omr_sheet.jpg","samples/community/Antibodyy/simple_omr_sheet.jpg","outputs/community/Antibodyy/CheckedOMRs/simple_omr_sheet.jpg","0","A","C","B","D","E","B"
+
+ ''',
+ })
+# ---
+# name: test_run_community_Sandeep_1507
+ dict({
+ 'Manual/ErrorFiles.csv': '''
+ "file_id","input_path","output_path","score","Booklet_No","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22","q23","q24","q25","q26","q27","q28","q29","q30","q31","q32","q33","q34","q35","q36","q37","q38","q39","q40","q41","q42","q43","q44","q45","q46","q47","q48","q49","q50","q51","q52","q53","q54","q55","q56","q57","q58","q59","q60","q61","q62","q63","q64","q65","q66","q67","q68","q69","q70","q71","q72","q73","q74","q75","q76","q77","q78","q79","q80","q81","q82","q83","q84","q85","q86","q87","q88","q89","q90","q91","q92","q93","q94","q95","q96","q97","q98","q99","q100","q101","q102","q103","q104","q105","q106","q107","q108","q109","q110","q111","q112","q113","q114","q115","q116","q117","q118","q119","q120","q121","q122","q123","q124","q125","q126","q127","q128","q129","q130","q131","q132","q133","q134","q135","q136","q137","q138","q139","q140","q141","q142","q143","q144","q145","q146","q147","q148","q149","q150","q151","q152","q153","q154","q155","q156","q157","q158","q159","q160","q161","q162","q163","q164","q165","q166","q167","q168","q169","q170","q171","q172","q173","q174","q175","q176","q177","q178","q179","q180","q181","q182","q183","q184","q185","q186","q187","q188","q189","q190","q191","q192","q193","q194","q195","q196","q197","q198","q199","q200"
+
+ ''',
+ 'Manual/MultiMarkedFiles.csv': '''
+ "file_id","input_path","output_path","score","Booklet_No","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22","q23","q24","q25","q26","q27","q28","q29","q30","q31","q32","q33","q34","q35","q36","q37","q38","q39","q40","q41","q42","q43","q44","q45","q46","q47","q48","q49","q50","q51","q52","q53","q54","q55","q56","q57","q58","q59","q60","q61","q62","q63","q64","q65","q66","q67","q68","q69","q70","q71","q72","q73","q74","q75","q76","q77","q78","q79","q80","q81","q82","q83","q84","q85","q86","q87","q88","q89","q90","q91","q92","q93","q94","q95","q96","q97","q98","q99","q100","q101","q102","q103","q104","q105","q106","q107","q108","q109","q110","q111","q112","q113","q114","q115","q116","q117","q118","q119","q120","q121","q122","q123","q124","q125","q126","q127","q128","q129","q130","q131","q132","q133","q134","q135","q136","q137","q138","q139","q140","q141","q142","q143","q144","q145","q146","q147","q148","q149","q150","q151","q152","q153","q154","q155","q156","q157","q158","q159","q160","q161","q162","q163","q164","q165","q166","q167","q168","q169","q170","q171","q172","q173","q174","q175","q176","q177","q178","q179","q180","q181","q182","q183","q184","q185","q186","q187","q188","q189","q190","q191","q192","q193","q194","q195","q196","q197","q198","q199","q200"
+
+ ''',
+ 'Results/Results_05AM.csv': '''
+ "file_id","input_path","output_path","score","Booklet_No","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22","q23","q24","q25","q26","q27","q28","q29","q30","q31","q32","q33","q34","q35","q36","q37","q38","q39","q40","q41","q42","q43","q44","q45","q46","q47","q48","q49","q50","q51","q52","q53","q54","q55","q56","q57","q58","q59","q60","q61","q62","q63","q64","q65","q66","q67","q68","q69","q70","q71","q72","q73","q74","q75","q76","q77","q78","q79","q80","q81","q82","q83","q84","q85","q86","q87","q88","q89","q90","q91","q92","q93","q94","q95","q96","q97","q98","q99","q100","q101","q102","q103","q104","q105","q106","q107","q108","q109","q110","q111","q112","q113","q114","q115","q116","q117","q118","q119","q120","q121","q122","q123","q124","q125","q126","q127","q128","q129","q130","q131","q132","q133","q134","q135","q136","q137","q138","q139","q140","q141","q142","q143","q144","q145","q146","q147","q148","q149","q150","q151","q152","q153","q154","q155","q156","q157","q158","q159","q160","q161","q162","q163","q164","q165","q166","q167","q168","q169","q170","q171","q172","q173","q174","q175","q176","q177","q178","q179","q180","q181","q182","q183","q184","q185","q186","q187","q188","q189","q190","q191","q192","q193","q194","q195","q196","q197","q198","q199","q200"
+ "omr-1.png","samples/community/Sandeep-1507/omr-1.png","outputs/community/Sandeep-1507/CheckedOMRs/omr-1.png","0","0190880","D","C","B","A","A","B","C","D","D","C","B","A","D","A","B","C","D","","B","D","C","A","C","C","B","A","D","A","AC","C","B","D","C","B","A","B","B","D","D","A","C","B","D","A","C","B","D","B","D","A","A","B","C","D","C","B","A","D","D","A","B","C","D","C","B","A","B","C","D","A","B","C","D","B","A","C","D","C","B","A","D","B","D","A","A","B","A","C","B","D","C","D","B","A","C","C","B","D","B","C","B","A","D","C","B","A","B","C","D","A","A","A","B","B","A","B","C","D","A","A","D","C","B","A","","A","B","C","D","D","D","B","B","C","C","D","C","C","D","D","C","C","B","B","A","A","D","D","B","A","D","C","B","A","A","D","D","B","B","A","A","B","C","D","D","C","B","A","B","D","A","C","C","C","A","A","B","B","D","D","A","A","B","C","D","B","D","A","B","C","D","AD","C","D","B","C","A","B","C","D"
+ "omr-2.png","samples/community/Sandeep-1507/omr-2.png","outputs/community/Sandeep-1507/CheckedOMRs/omr-2.png","0","0no22nonono","A","B","B","A","D","C","B","D","C","D","D","D","B","B","D","D","D","B","C","C","A","A","B","A","D","A","A","B","A","C","A","C","D","D","D","","","C","C","B","B","B","","D","","C","D","","D","B","A","D","B","A","C","A","C","A","C","B","A","D","C","B","C","B","C","D","B","B","D","C","C","D","D","A","D","A","D","C","B","D","C","A","C","","C","B","B","","A","A","D","","B","A","","C","A","D","D","C","C","A","C","A","C","D","A","A","A","D","D","B","C","B","B","B","D","A","C","D","D","A","A","A","C","D","C","C","B","D","A","A","C","B","","D","A","C","C","C","","","","A","C","","D","A","B","A","A","C","A","D","B","B","A","D","A","B","C","A","C","D","D","D","C","A","C","A","C","D","A","A","A","D","A","B","A","B","C","B","A","","B","C","D","D","","","D","C","C","C","","C","A",""
+ "omr-3.png","samples/community/Sandeep-1507/omr-3.png","outputs/community/Sandeep-1507/CheckedOMRs/omr-3.png","0","0nononono73","B","A","C","D","A","D","D","A","C","A","A","B","C","A","A","C","A","B","A","D","C","C","A","D","D","C","C","C","A","C","C","B","B","D","D","C","","","C","B","","","D","A","A","A","A","","A","C","C","C","D","C","","A","B","C","D","B","C","C","C","D","A","B","B","B","D","D","B","B","C","D","B","D","A","B","A","B","C","A","C","A","C","D","","","A","B","","B","C","D","A","D","D","","","C","D","B","B","A","A","D","D","B","A","B","B","C","C","D","D","C","A","D","C","D","C","C","B","C","D","C","D","A","B","D","C","B","D","B","B","","D","","B","D","B","B","C","A","D","","C","","C","","B","C","A","B","B","D","D","D","B","A","D","D","A","D","D","C","B","B","D","C","B","A","C","D","A","D","D","A","C","A","B","D","C","C","C","A","D","","","B","B","","C","C","B","B","C","","","B"
+
+ ''',
+ })
+# ---
+# name: test_run_community_Shamanth
+ dict({
+ 'Manual/ErrorFiles.csv': '''
+ "file_id","input_path","output_path","score","q21","q22","q23","q24","q25","q26","q27","q28"
+
+ ''',
+ 'Manual/MultiMarkedFiles.csv': '''
+ "file_id","input_path","output_path","score","q21","q22","q23","q24","q25","q26","q27","q28"
+
+ ''',
+ 'Results/Results_05AM.csv': '''
+ "file_id","input_path","output_path","score","q21","q22","q23","q24","q25","q26","q27","q28"
+ "omr_sheet_01.png","samples/community/Shamanth/omr_sheet_01.png","outputs/community/Shamanth/CheckedOMRs/omr_sheet_01.png","0","A","B","C","D","A","C","C","D"
+
+ ''',
+ })
+# ---
+# name: test_run_community_UPSC_mock
+ dict({
+ 'Manual/ErrorFiles.csv': '''
+ "file_id","input_path","output_path","score","Roll","Subject Code","bookletNo","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22","q23","q24","q25","q26","q27","q28","q29","q30","q31","q32","q33","q34","q35","q36","q37","q38","q39","q40","q41","q42","q43","q44","q45","q46","q47","q48","q49","q50","q51","q52","q53","q54","q55","q56","q57","q58","q59","q60","q61","q62","q63","q64","q65","q66","q67","q68","q69","q70","q71","q72","q73","q74","q75","q76","q77","q78","q79","q80","q81","q82","q83","q84","q85","q86","q87","q88","q89","q90","q91","q92","q93","q94","q95","q96","q97","q98","q99","q100"
+
+ ''',
+ 'Manual/MultiMarkedFiles.csv': '''
+ "file_id","input_path","output_path","score","Roll","Subject Code","bookletNo","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22","q23","q24","q25","q26","q27","q28","q29","q30","q31","q32","q33","q34","q35","q36","q37","q38","q39","q40","q41","q42","q43","q44","q45","q46","q47","q48","q49","q50","q51","q52","q53","q54","q55","q56","q57","q58","q59","q60","q61","q62","q63","q64","q65","q66","q67","q68","q69","q70","q71","q72","q73","q74","q75","q76","q77","q78","q79","q80","q81","q82","q83","q84","q85","q86","q87","q88","q89","q90","q91","q92","q93","q94","q95","q96","q97","q98","q99","q100"
+
+ ''',
+ 'Results/Results_05AM.csv': '''
+ "file_id","input_path","output_path","score","Roll","Subject Code","bookletNo","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22","q23","q24","q25","q26","q27","q28","q29","q30","q31","q32","q33","q34","q35","q36","q37","q38","q39","q40","q41","q42","q43","q44","q45","q46","q47","q48","q49","q50","q51","q52","q53","q54","q55","q56","q57","q58","q59","q60","q61","q62","q63","q64","q65","q66","q67","q68","q69","q70","q71","q72","q73","q74","q75","q76","q77","q78","q79","q80","q81","q82","q83","q84","q85","q86","q87","q88","q89","q90","q91","q92","q93","q94","q95","q96","q97","q98","q99","q100"
+ "answer_key.jpg","samples/community/UPSC-mock/answer_key.jpg","outputs/community/UPSC-mock/CheckedOMRs/answer_key.jpg","200.0","","","","C","D","A","C","C","C","B","A","C","C","B","D","B","D","C","C","B","D","B","D","C","C","C","B","D","D","D","B","A","D","D","C","A","B","C","A","D","A","A","A","D","D","B","A","B","C","B","A","C","D","C","D","A","B","C","A","C","C","C","D","B","C","C","C","C","A","D","A","D","A","D","C","C","D","C","D","A","A","C","B","C","D","C","A","B","C","B","D","A","A","C","A","B","D","C","D","A","C","B","A"
+
+ ''',
+ 'scan-angles/Manual/ErrorFiles.csv': '''
+ "file_id","input_path","output_path","score","Roll","Subject Code","bookletNo","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22","q23","q24","q25","q26","q27","q28","q29","q30","q31","q32","q33","q34","q35","q36","q37","q38","q39","q40","q41","q42","q43","q44","q45","q46","q47","q48","q49","q50","q51","q52","q53","q54","q55","q56","q57","q58","q59","q60","q61","q62","q63","q64","q65","q66","q67","q68","q69","q70","q71","q72","q73","q74","q75","q76","q77","q78","q79","q80","q81","q82","q83","q84","q85","q86","q87","q88","q89","q90","q91","q92","q93","q94","q95","q96","q97","q98","q99","q100"
+
+ ''',
+ 'scan-angles/Manual/MultiMarkedFiles.csv': '''
+ "file_id","input_path","output_path","score","Roll","Subject Code","bookletNo","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22","q23","q24","q25","q26","q27","q28","q29","q30","q31","q32","q33","q34","q35","q36","q37","q38","q39","q40","q41","q42","q43","q44","q45","q46","q47","q48","q49","q50","q51","q52","q53","q54","q55","q56","q57","q58","q59","q60","q61","q62","q63","q64","q65","q66","q67","q68","q69","q70","q71","q72","q73","q74","q75","q76","q77","q78","q79","q80","q81","q82","q83","q84","q85","q86","q87","q88","q89","q90","q91","q92","q93","q94","q95","q96","q97","q98","q99","q100"
+
+ ''',
+ 'scan-angles/Results/Results_05AM.csv': '''
+ "file_id","input_path","output_path","score","Roll","Subject Code","bookletNo","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22","q23","q24","q25","q26","q27","q28","q29","q30","q31","q32","q33","q34","q35","q36","q37","q38","q39","q40","q41","q42","q43","q44","q45","q46","q47","q48","q49","q50","q51","q52","q53","q54","q55","q56","q57","q58","q59","q60","q61","q62","q63","q64","q65","q66","q67","q68","q69","q70","q71","q72","q73","q74","q75","q76","q77","q78","q79","q80","q81","q82","q83","q84","q85","q86","q87","q88","q89","q90","q91","q92","q93","q94","q95","q96","q97","q98","q99","q100"
+ "angle-1.jpg","samples/community/UPSC-mock/scan-angles/angle-1.jpg","outputs/community/UPSC-mock/scan-angles/CheckedOMRs/angle-1.jpg","70.66666666666669","","","","D","D","A","","C","C","B","","A","C","C","D","A","D","A","C","A","D","B","D","D","C","D","D","D","D","","B","A","D","D","C","","B","","C","D","","","A","","A","C","C","B","C","A","A","C","","C","","D","B","C","","B","C","D","","","C","C","","C","A","B","C","","","","","D","D","C","D","A","","","B","","B","D","C","C","","D","","D","C","D","A","","A","","","A","C","B","A"
+ "angle-2.jpg","samples/community/UPSC-mock/scan-angles/angle-2.jpg","outputs/community/UPSC-mock/scan-angles/CheckedOMRs/angle-2.jpg","70.66666666666669","","","","D","D","A","","C","C","B","","A","C","C","D","A","D","A","C","A","D","B","D","D","C","D","D","D","D","","B","A","D","D","C","","B","","C","D","","","A","","A","C","C","B","C","A","A","C","","C","","D","B","C","","B","C","D","","","C","C","","C","A","B","C","","","","","D","D","C","D","A","","","B","","B","D","C","C","","D","","D","C","D","A","","A","","","A","C","B","A"
+ "angle-3.jpg","samples/community/UPSC-mock/scan-angles/angle-3.jpg","outputs/community/UPSC-mock/scan-angles/CheckedOMRs/angle-3.jpg","70.66666666666669","","","","D","D","A","","C","C","B","","A","C","C","D","A","D","A","C","A","D","B","D","D","C","D","D","D","D","","B","A","D","D","C","","B","","C","D","","","A","","A","C","C","B","C","A","A","C","","C","","D","B","C","","B","C","D","","","C","C","","C","A","B","C","","","","","D","D","C","D","A","","","B","","B","D","C","C","","D","","D","C","D","A","","A","","","A","C","B","A"
+
+ ''',
+ })
+# ---
+# name: test_run_community_UmarFarootAPS
+ dict({
+ 'scans/Manual/ErrorFiles.csv': '''
+ "file_id","input_path","output_path","score","Roll_no","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22","q23","q24","q25","q26","q27","q28","q29","q30","q31","q32","q33","q34","q35","q36","q37","q38","q39","q40","q41","q42","q43","q44","q45","q46","q47","q48","q49","q50","q51","q52","q53","q54","q55","q56","q57","q58","q59","q60","q61","q62","q63","q64","q65","q66","q67","q68","q69","q70","q71","q72","q73","q74","q75","q76","q77","q78","q79","q80","q81","q82","q83","q84","q85","q86","q87","q88","q89","q90","q91","q92","q93","q94","q95","q96","q97","q98","q99","q100","q101","q102","q103","q104","q105","q106","q107","q108","q109","q110","q111","q112","q113","q114","q115","q116","q117","q118","q119","q120","q121","q122","q123","q124","q125","q126","q127","q128","q129","q130","q131","q132","q133","q134","q135","q136","q137","q138","q139","q140","q141","q142","q143","q144","q145","q146","q147","q148","q149","q150","q151","q152","q153","q154","q155","q156","q157","q158","q159","q160","q161","q162","q163","q164","q165","q166","q167","q168","q169","q170","q171","q172","q173","q174","q175","q176","q177","q178","q179","q180","q181","q182","q183","q184","q185","q186","q187","q188","q189","q190","q191","q192","q193","q194","q195","q196","q197","q198","q199","q200"
+
+ ''',
+ 'scans/Manual/MultiMarkedFiles.csv': '''
+ "file_id","input_path","output_path","score","Roll_no","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22","q23","q24","q25","q26","q27","q28","q29","q30","q31","q32","q33","q34","q35","q36","q37","q38","q39","q40","q41","q42","q43","q44","q45","q46","q47","q48","q49","q50","q51","q52","q53","q54","q55","q56","q57","q58","q59","q60","q61","q62","q63","q64","q65","q66","q67","q68","q69","q70","q71","q72","q73","q74","q75","q76","q77","q78","q79","q80","q81","q82","q83","q84","q85","q86","q87","q88","q89","q90","q91","q92","q93","q94","q95","q96","q97","q98","q99","q100","q101","q102","q103","q104","q105","q106","q107","q108","q109","q110","q111","q112","q113","q114","q115","q116","q117","q118","q119","q120","q121","q122","q123","q124","q125","q126","q127","q128","q129","q130","q131","q132","q133","q134","q135","q136","q137","q138","q139","q140","q141","q142","q143","q144","q145","q146","q147","q148","q149","q150","q151","q152","q153","q154","q155","q156","q157","q158","q159","q160","q161","q162","q163","q164","q165","q166","q167","q168","q169","q170","q171","q172","q173","q174","q175","q176","q177","q178","q179","q180","q181","q182","q183","q184","q185","q186","q187","q188","q189","q190","q191","q192","q193","q194","q195","q196","q197","q198","q199","q200"
+
+ ''',
+ 'scans/Results/Results_05AM.csv': '''
+ "file_id","input_path","output_path","score","Roll_no","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22","q23","q24","q25","q26","q27","q28","q29","q30","q31","q32","q33","q34","q35","q36","q37","q38","q39","q40","q41","q42","q43","q44","q45","q46","q47","q48","q49","q50","q51","q52","q53","q54","q55","q56","q57","q58","q59","q60","q61","q62","q63","q64","q65","q66","q67","q68","q69","q70","q71","q72","q73","q74","q75","q76","q77","q78","q79","q80","q81","q82","q83","q84","q85","q86","q87","q88","q89","q90","q91","q92","q93","q94","q95","q96","q97","q98","q99","q100","q101","q102","q103","q104","q105","q106","q107","q108","q109","q110","q111","q112","q113","q114","q115","q116","q117","q118","q119","q120","q121","q122","q123","q124","q125","q126","q127","q128","q129","q130","q131","q132","q133","q134","q135","q136","q137","q138","q139","q140","q141","q142","q143","q144","q145","q146","q147","q148","q149","q150","q151","q152","q153","q154","q155","q156","q157","q158","q159","q160","q161","q162","q163","q164","q165","q166","q167","q168","q169","q170","q171","q172","q173","q174","q175","q176","q177","q178","q179","q180","q181","q182","q183","q184","q185","q186","q187","q188","q189","q190","q191","q192","q193","q194","q195","q196","q197","q198","q199","q200"
+ "scan-type-1.jpg","samples/community/UmarFarootAPS/scans/scan-type-1.jpg","outputs/community/UmarFarootAPS/scans/CheckedOMRs/scan-type-1.jpg","49.0","2468","A","C","B","C","A","D","B","C","B","D","C","A","C","D","B","C","A","B","C","A","C","B","D","C","A","B","D","C","A","C","B","D","B","A","C","D","B","C","A","C","D","A","C","D","A","B","D","C","A","C","D","B","C","A","C","D","B","C","D","A","B","C","B","C","D","B","D","A","C","B","D","A","B","C","B","A","C","D","B","A","C","B","C","B","A","D","B","A","C","D","B","D","B","C","B","D","A","C","B","C","B","C","D","B","C","A","B","C","A","D","C","B","D","B","A","B","C","D","D","C","B","A","B","C","D","C","B","A","B","C","D","C","B","A","B","C","D","C","B","A","B","C","B","A","C","B","A","C","A","B","C","B","C","B","A","C","A","C","B","B","C","B","A","C","A","B","A","B","A","B","C","D","B","C","A","C","D","C","A","C","B","A","C","A","B","C","B","D","A","B","C","D","C","B","B","C","A","B","C","B"
+ "scan-type-2.jpg","samples/community/UmarFarootAPS/scans/scan-type-2.jpg","outputs/community/UmarFarootAPS/scans/CheckedOMRs/scan-type-2.jpg","20.0","0234","A","B","C","D","C","B","A","B","C","D","C","B","A","B","C","D","C","B","A","B","C","D","C","B","A","B","C","D","C","B","A","B","C","D","C","B","A","B","C","D","C","B","A","B","C","D","C","B","A","B","A","D","","","AD","","","","A","D","","","","","","","D","A","","D","","A","","D","","","","A","","","C","","","D","","","A","","","","D","","C","","A","","C","","D","B","B","","","A","","D","","","","D","","","","","A","D","","","B","","","D","","","A","","","D","","","","","","D","","","","A","D","","","A","","B","","D","","","","C","C","D","D","A","","D","","A","D","","","D","","B","D","","","D","","D","B","","","","D","","A","","","","D","","B","","","","","","D","","","A","","","A","","D","","","D"
+
+ ''',
+ })
+# ---
+# name: test_run_community_ibrahimkilic
+ dict({
+ 'Manual/ErrorFiles.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5"
+
+ ''',
+ 'Manual/MultiMarkedFiles.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5"
+
+ ''',
+ 'Results/Results_05AM.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5"
+ "yes_no_questionnarie.jpg","samples/community/ibrahimkilic/yes_no_questionnarie.jpg","outputs/community/ibrahimkilic/CheckedOMRs/yes_no_questionnarie.jpg","0","no","no","no","no","no"
+
+ ''',
+ })
+# ---
+# name: test_run_sample1
+ dict({
+ 'MobileCamera/Manual/ErrorFiles.csv': '''
+ "file_id","input_path","output_path","score","Roll","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20"
+
+ ''',
+ 'MobileCamera/Manual/MultiMarkedFiles.csv': '''
+ "file_id","input_path","output_path","score","Roll","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20"
+
+ ''',
+ 'MobileCamera/Results/Results_05AM.csv': '''
+ "file_id","input_path","output_path","score","Roll","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20"
+ "sheet1.jpg","samples/sample1/MobileCamera/sheet1.jpg","outputs/sample1/MobileCamera/CheckedOMRs/sheet1.jpg","0","E503110026","B","","D","B","6","11","20","7","16","B","D","C","D","A","D","B","A","C","C","D"
+
+ ''',
+ })
+# ---
+# name: test_run_sample2
+ dict({
+ 'AdrianSample/Manual/ErrorFiles.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5"
+
+ ''',
+ 'AdrianSample/Manual/MultiMarkedFiles.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5"
+
+ ''',
+ 'AdrianSample/Results/Results_05AM.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5"
+ "adrian_omr.png","samples/sample2/AdrianSample/adrian_omr.png","outputs/sample2/AdrianSample/CheckedOMRs/adrian_omr.png","0","B","E","A","C","B"
+ "adrian_omr_2.png","samples/sample2/AdrianSample/adrian_omr_2.png","outputs/sample2/AdrianSample/CheckedOMRs/adrian_omr_2.png","0","C","E","A","B","B"
+
+ ''',
+ })
+# ---
+# name: test_run_sample3
+ dict({
+ 'colored-thick-sheet/Manual/ErrorFiles.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22","q23","q24","q25","q26","q27","q28","q29","q30","q31","q32","q33","q34","q35","q36","q37","q38","q39","q40","q41","q42","q43","q44","q45","q46","q47","q48","q49","q50","q51","q52","q53","q54","q55","q56","q57","q58","q59","q60","q61","q62","q63","q64","q65","q66","q67","q68","q69","q70","q71","q72","q73","q74","q75","q76","q77","q78","q79","q80","q81","q82","q83","q84","q85","q86","q87","q88","q89","q90","q91","q92","q93","q94","q95","q96","q97","q98","q99","q100"
+
+ ''',
+ 'colored-thick-sheet/Manual/MultiMarkedFiles.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22","q23","q24","q25","q26","q27","q28","q29","q30","q31","q32","q33","q34","q35","q36","q37","q38","q39","q40","q41","q42","q43","q44","q45","q46","q47","q48","q49","q50","q51","q52","q53","q54","q55","q56","q57","q58","q59","q60","q61","q62","q63","q64","q65","q66","q67","q68","q69","q70","q71","q72","q73","q74","q75","q76","q77","q78","q79","q80","q81","q82","q83","q84","q85","q86","q87","q88","q89","q90","q91","q92","q93","q94","q95","q96","q97","q98","q99","q100"
+
+ ''',
+ 'colored-thick-sheet/Results/Results_05AM.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22","q23","q24","q25","q26","q27","q28","q29","q30","q31","q32","q33","q34","q35","q36","q37","q38","q39","q40","q41","q42","q43","q44","q45","q46","q47","q48","q49","q50","q51","q52","q53","q54","q55","q56","q57","q58","q59","q60","q61","q62","q63","q64","q65","q66","q67","q68","q69","q70","q71","q72","q73","q74","q75","q76","q77","q78","q79","q80","q81","q82","q83","q84","q85","q86","q87","q88","q89","q90","q91","q92","q93","q94","q95","q96","q97","q98","q99","q100"
+ "rgb-100-gsm.jpg","samples/sample3/colored-thick-sheet/rgb-100-gsm.jpg","outputs/sample3/colored-thick-sheet/CheckedOMRs/rgb-100-gsm.jpg","0","D","D","A","","C","C","B","","A","C","C","D","A","D","A","C","A","D","B","D","D","C","D","D","D","D","","B","A","D","D","C","","B","","C","D","","","A","","A","C","C","B","C","A","A","C","","C","","D","B","C","","B","C","D","","","C","C","","C","A","B","C","","","","","D","D","C","D","A","","","B","","B","D","C","C","","D","","D","C","D","A","","A","","","A","C","B","A"
+
+ ''',
+ 'xeroxed-thin-sheet/Manual/ErrorFiles.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22","q23","q24","q25","q26","q27","q28","q29","q30","q31","q32","q33","q34","q35","q36","q37","q38","q39","q40","q41","q42","q43","q44","q45","q46","q47","q48","q49","q50","q51","q52","q53","q54","q55","q56","q57","q58","q59","q60","q61","q62","q63","q64","q65","q66","q67","q68","q69","q70","q71","q72","q73","q74","q75","q76","q77","q78","q79","q80","q81","q82","q83","q84","q85","q86","q87","q88","q89","q90","q91","q92","q93","q94","q95","q96","q97","q98","q99","q100"
+
+ ''',
+ 'xeroxed-thin-sheet/Manual/MultiMarkedFiles.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22","q23","q24","q25","q26","q27","q28","q29","q30","q31","q32","q33","q34","q35","q36","q37","q38","q39","q40","q41","q42","q43","q44","q45","q46","q47","q48","q49","q50","q51","q52","q53","q54","q55","q56","q57","q58","q59","q60","q61","q62","q63","q64","q65","q66","q67","q68","q69","q70","q71","q72","q73","q74","q75","q76","q77","q78","q79","q80","q81","q82","q83","q84","q85","q86","q87","q88","q89","q90","q91","q92","q93","q94","q95","q96","q97","q98","q99","q100"
+
+ ''',
+ 'xeroxed-thin-sheet/Results/Results_05AM.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22","q23","q24","q25","q26","q27","q28","q29","q30","q31","q32","q33","q34","q35","q36","q37","q38","q39","q40","q41","q42","q43","q44","q45","q46","q47","q48","q49","q50","q51","q52","q53","q54","q55","q56","q57","q58","q59","q60","q61","q62","q63","q64","q65","q66","q67","q68","q69","q70","q71","q72","q73","q74","q75","q76","q77","q78","q79","q80","q81","q82","q83","q84","q85","q86","q87","q88","q89","q90","q91","q92","q93","q94","q95","q96","q97","q98","q99","q100"
+ "grayscale-80-gsm.jpg","samples/sample3/xeroxed-thin-sheet/grayscale-80-gsm.jpg","outputs/sample3/xeroxed-thin-sheet/CheckedOMRs/grayscale-80-gsm.jpg","0","C","D","A","C","C","C","B","A","C","C","B","D","B","D","C","C","B","D","B","D","C","C","C","B","D","D","D","B","A","D","D","C","A","B","C","A","D","A","A","A","D","D","B","A","B","C","B","A","C","D","C","D","A","B","C","A","C","C","C","D","B","C","C","C","C","A","D","A","D","A","D","C","C","D","C","D","A","A","C","B","C","D","C","A","B","C","B","D","A","A","C","A","B","D","C","D","A","C","B","A"
+
+ ''',
+ })
+# ---
+# name: test_run_sample4
+ dict({
+ 'Manual/ErrorFiles.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11"
+
+ ''',
+ 'Manual/MultiMarkedFiles.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11"
+
+ ''',
+ 'Results/Results_05AM.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11"
+ "IMG_20201116_143512.jpg","samples/sample4/IMG_20201116_143512.jpg","outputs/sample4/CheckedOMRs/IMG_20201116_143512.jpg","33.0","B","D","C","B","D","C","BC","A","C","D","C"
+ "IMG_20201116_150717658.jpg","samples/sample4/IMG_20201116_150717658.jpg","outputs/sample4/CheckedOMRs/IMG_20201116_150717658.jpg","33.0","B","D","C","B","D","C","BC","A","C","D","C"
+ "IMG_20201116_150750830.jpg","samples/sample4/IMG_20201116_150750830.jpg","outputs/sample4/CheckedOMRs/IMG_20201116_150750830.jpg","-2.0","A","","D","C","AC","A","D","B","C","D","D"
+
+ ''',
+ })
+# ---
+# name: test_run_sample5
+ dict({
+ 'ScanBatch1/Manual/ErrorFiles.csv': '''
+ "file_id","input_path","output_path","score","Roll","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22"
+
+ ''',
+ 'ScanBatch1/Manual/MultiMarkedFiles.csv': '''
+ "file_id","input_path","output_path","score","Roll","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22"
+
+ ''',
+ 'ScanBatch1/Results/Results_05AM.csv': '''
+ "file_id","input_path","output_path","score","Roll","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22"
+ "camscanner-1.jpg","samples/sample5/ScanBatch1/camscanner-1.jpg","outputs/sample5/ScanBatch1/CheckedOMRs/camscanner-1.jpg","-4.0","E204420102","D","C","A","C","B","08","52","21","85","36","B","C","A","A","D","C","C","AD","A","A","D",""
+
+ ''',
+ 'ScanBatch2/Manual/ErrorFiles.csv': '''
+ "file_id","input_path","output_path","score","Roll","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22"
+
+ ''',
+ 'ScanBatch2/Manual/MultiMarkedFiles.csv': '''
+ "file_id","input_path","output_path","score","Roll","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22"
+
+ ''',
+ 'ScanBatch2/Results/Results_05AM.csv': '''
+ "file_id","input_path","output_path","score","Roll","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22"
+ "camscanner-2.jpg","samples/sample5/ScanBatch2/camscanner-2.jpg","outputs/sample5/ScanBatch2/CheckedOMRs/camscanner-2.jpg","55.0","E204420109","C","C","B","C","C","01","19","10","10","18","D","A","D","D","D","C","C","C","C","D","B","A"
+
+ ''',
+ })
+# ---
+# name: test_run_sample6
+ dict({
+ 'Manual/ErrorFiles.csv': '''
+ "file_id","input_path","output_path","score","Roll"
+
+ ''',
+ 'Manual/MultiMarkedFiles.csv': '''
+ "file_id","input_path","output_path","score","Roll"
+
+ ''',
+ 'Results/Results_05AM.csv': '''
+ "file_id","input_path","output_path","score","Roll"
+ "reference.png","samples/sample6/reference.png","outputs/sample6/CheckedOMRs/reference.png","0","A"
+
+ ''',
+ 'doc-scans/Manual/ErrorFiles.csv': '''
+ "file_id","input_path","output_path","score","Roll"
+
+ ''',
+ 'doc-scans/Manual/MultiMarkedFiles.csv': '''
+ "file_id","input_path","output_path","score","Roll"
+
+ ''',
+ 'doc-scans/Results/Results_05AM.csv': '''
+ "file_id","input_path","output_path","score","Roll"
+ "sample_roll_01.jpg","samples/sample6/doc-scans/sample_roll_01.jpg","outputs/sample6/doc-scans/CheckedOMRs/sample_roll_01.jpg","0","A0188877Y"
+ "sample_roll_02.jpg","samples/sample6/doc-scans/sample_roll_02.jpg","outputs/sample6/doc-scans/CheckedOMRs/sample_roll_02.jpg","0","A0203959W"
+ "sample_roll_03.jpg","samples/sample6/doc-scans/sample_roll_03.jpg","outputs/sample6/doc-scans/CheckedOMRs/sample_roll_03.jpg","0","A0204729A"
+
+ ''',
+ })
+# ---
diff --git a/src/tests/test_all_samples.py b/src/tests/test_all_samples.py
new file mode 100644
index 00000000..9efe94ff
--- /dev/null
+++ b/src/tests/test_all_samples.py
@@ -0,0 +1,112 @@
+import os
+import shutil
+from glob import glob
+
+from src.tests.utils import run_entry_point, setup_mocker_patches
+
+
+def read_file(path):
+ with open(path) as file:
+ return file.read()
+
+
+def run_sample(mocker, sample_path):
+ setup_mocker_patches(mocker)
+
+ input_path = os.path.join("samples", sample_path)
+ output_dir = os.path.join("outputs", sample_path)
+ if os.path.exists(output_dir):
+ print(
+ f"Warning: output directory already exists: {output_dir}. This may affect the test execution."
+ )
+
+ run_entry_point(input_path, output_dir)
+
+ sample_outputs = extract_sample_outputs(output_dir)
+
+ print(f"Note: removing output directory: {output_dir}")
+ shutil.rmtree(output_dir)
+
+ return sample_outputs
+
+
+EXT = "*.csv"
+
+
+def extract_sample_outputs(output_dir):
+ sample_outputs = {}
+ for _dir, _subdir, _files in os.walk(output_dir):
+ for file in glob(os.path.join(_dir, EXT)):
+ relative_path = os.path.relpath(file, output_dir)
+ sample_outputs[relative_path] = read_file(file)
+ return sample_outputs
+
+
+def test_run_answer_key_using_csv(mocker, snapshot):
+ sample_outputs = run_sample(mocker, "answer-key/using-csv")
+ assert snapshot == sample_outputs
+
+
+def test_run_answer_key_weighted_answers(mocker, snapshot):
+ sample_outputs = run_sample(mocker, "answer-key/weighted-answers")
+ assert snapshot == sample_outputs
+
+
+def test_run_sample1(mocker, snapshot):
+ sample_outputs = run_sample(mocker, "sample1")
+ assert snapshot == sample_outputs
+
+
+def test_run_sample2(mocker, snapshot):
+ sample_outputs = run_sample(mocker, "sample2")
+ assert snapshot == sample_outputs
+
+
+def test_run_sample3(mocker, snapshot):
+ sample_outputs = run_sample(mocker, "sample3")
+ assert snapshot == sample_outputs
+
+
+def test_run_sample4(mocker, snapshot):
+ sample_outputs = run_sample(mocker, "sample4")
+ assert snapshot == sample_outputs
+
+
+def test_run_sample5(mocker, snapshot):
+ sample_outputs = run_sample(mocker, "sample5")
+ assert snapshot == sample_outputs
+
+
+def test_run_sample6(mocker, snapshot):
+ sample_outputs = run_sample(mocker, "sample6")
+ assert snapshot == sample_outputs
+
+
+def test_run_community_Antibodyy(mocker, snapshot):
+ sample_outputs = run_sample(mocker, "community/Antibodyy")
+ assert snapshot == sample_outputs
+
+
+def test_run_community_ibrahimkilic(mocker, snapshot):
+ sample_outputs = run_sample(mocker, "community/ibrahimkilic")
+ assert snapshot == sample_outputs
+
+
+def test_run_community_Sandeep_1507(mocker, snapshot):
+ sample_outputs = run_sample(mocker, "community/Sandeep-1507")
+ assert snapshot == sample_outputs
+
+
+def test_run_community_Shamanth(mocker, snapshot):
+ sample_outputs = run_sample(mocker, "community/Shamanth")
+ assert snapshot == sample_outputs
+
+
+def test_run_community_UmarFarootAPS(mocker, snapshot):
+ sample_outputs = run_sample(mocker, "community/UmarFarootAPS")
+ assert snapshot == sample_outputs
+
+
+def test_run_community_UPSC_mock(mocker, snapshot):
+ sample_outputs = run_sample(mocker, "community/UPSC-mock")
+ assert snapshot == sample_outputs
diff --git a/src/tests/test_edge_cases.py b/src/tests/test_edge_cases.py
new file mode 100644
index 00000000..f3d6fd45
--- /dev/null
+++ b/src/tests/test_edge_cases.py
@@ -0,0 +1,95 @@
+import os
+from pathlib import Path
+
+import pandas as pd
+
+from src.tests.test_samples.sample2.boilerplate import (
+ CONFIG_BOILERPLATE,
+ TEMPLATE_BOILERPLATE,
+)
+from src.tests.utils import (
+ generate_write_jsons_and_run,
+ remove_file,
+ run_entry_point,
+ setup_mocker_patches,
+)
+
+FROZEN_TIMESTAMP = "1970-01-01"
+CURRENT_DIR = Path("src/tests")
+BASE_SAMPLE_PATH = CURRENT_DIR.joinpath("test_samples", "sample2")
+BASE_RESULTS_CSV_PATH = os.path.join(
+ "outputs", BASE_SAMPLE_PATH, "Results", "Results_05AM.csv"
+)
+BASE_MULTIMARKED_CSV_PATH = os.path.join(
+ "outputs", BASE_SAMPLE_PATH, "Manual", "MultiMarkedFiles.csv"
+)
+
+
+def run_sample(mocker, input_path):
+ setup_mocker_patches(mocker)
+ output_dir = os.path.join("outputs", input_path)
+ run_entry_point(input_path, output_dir)
+
+
+def extract_output_data(path):
+ output_data = pd.read_csv(path, keep_default_na=False)
+ return output_data
+
+
+write_jsons_and_run = generate_write_jsons_and_run(
+ run_sample,
+ sample_path=BASE_SAMPLE_PATH,
+ template_boilerplate=TEMPLATE_BOILERPLATE,
+ config_boilerplate=CONFIG_BOILERPLATE,
+)
+
+
+def test_config_low_dimensions(mocker):
+ def modify_config(config):
+ config["dimensions"]["processing_height"] = 1000
+ config["dimensions"]["processing_width"] = 1000
+
+ exception = write_jsons_and_run(mocker, modify_config=modify_config)
+
+ assert str(exception) == "No Error"
+
+
+def test_different_bubble_dimensions(mocker):
+ # Prevent appending to output csv:
+ remove_file(BASE_RESULTS_CSV_PATH)
+ remove_file(BASE_MULTIMARKED_CSV_PATH)
+
+ exception = write_jsons_and_run(mocker)
+ assert str(exception) == "No Error"
+ original_output_data = extract_output_data(BASE_RESULTS_CSV_PATH)
+
+ def modify_template(template):
+ # Incorrect global bubble size
+ template["bubbleDimensions"] = [5, 5]
+ # Correct bubble size for MCQBlock1a1
+ template["fieldBlocks"]["MCQBlock1a1"]["bubbleDimensions"] = [32, 32]
+ # Incorrect bubble size for MCQBlock1a11
+ template["fieldBlocks"]["MCQBlock1a11"]["bubbleDimensions"] = [10, 10]
+
+ remove_file(BASE_RESULTS_CSV_PATH)
+ remove_file(BASE_MULTIMARKED_CSV_PATH)
+ exception = write_jsons_and_run(mocker, modify_template=modify_template)
+ assert str(exception) == "No Error"
+
+ results_output_data = extract_output_data(BASE_RESULTS_CSV_PATH)
+
+ assert results_output_data.empty
+
+ output_data = extract_output_data(BASE_MULTIMARKED_CSV_PATH)
+
+ equal_columns = [f"q{i}" for i in range(1, 18)]
+ assert (
+ output_data[equal_columns].iloc[0].to_list()
+ == original_output_data[equal_columns].iloc[0].to_list()
+ )
+
+ unequal_columns = [f"q{i}" for i in range(168, 185)]
+ assert not (
+ output_data[unequal_columns].iloc[0].to_list()
+ == original_output_data[unequal_columns].iloc[0].to_list()
+ )
diff --git a/src/tests/test_samples/sample1/boilerplate.py b/src/tests/test_samples/sample1/boilerplate.py
new file mode 100644
index 00000000..74dda465
--- /dev/null
+++ b/src/tests/test_samples/sample1/boilerplate.py
@@ -0,0 +1,14 @@
+TEMPLATE_BOILERPLATE = {
+ "pageDimensions": [300, 400],
+ "bubbleDimensions": [25, 25],
+ "preProcessors": [{"name": "CropPage", "options": {"morphKernel": [10, 10]}}],
+ "fieldBlocks": {
+ "MCQ_Block_1": {
+ "fieldType": "QTYPE_MCQ5",
+ "origin": [65, 60],
+ "fieldLabels": ["q1..5"],
+ "labelsGap": 52,
+ "bubblesGap": 41,
+ }
+ },
+}
diff --git a/src/tests/test_samples/sample1/sample.png b/src/tests/test_samples/sample1/sample.png
new file mode 100644
index 00000000..d8db0994
Binary files /dev/null and b/src/tests/test_samples/sample1/sample.png differ
diff --git a/src/tests/test_samples/sample2/boilerplate.py b/src/tests/test_samples/sample2/boilerplate.py
new file mode 100644
index 00000000..f43c3093
--- /dev/null
+++ b/src/tests/test_samples/sample2/boilerplate.py
@@ -0,0 +1,39 @@
+TEMPLATE_BOILERPLATE = {
+ "pageDimensions": [2550, 3300],
+ "bubbleDimensions": [32, 32],
+ "preProcessors": [
+ {
+ "name": "CropOnMarkers",
+ "options": {
+ "relativePath": "omr_marker.jpg",
+ "sheetToMarkerWidthRatio": 17,
+ },
+ }
+ ],
+ "fieldBlocks": {
+ "MCQBlock1a1": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [197, 300],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": ["q1..17"],
+ },
+ "MCQBlock1a11": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [1770, 1310],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": ["q168..184"],
+ },
+ },
+}
+
+CONFIG_BOILERPLATE = {
+ "dimensions": {
+ "display_height": 960,
+ "display_width": 1280,
+ "processing_height": 1640,
+ "processing_width": 1332,
+ },
+ "outputs": {"show_image_level": 0, "filter_out_multimarked_files": True},
+}
diff --git a/src/tests/test_samples/sample2/omr_marker.jpg b/src/tests/test_samples/sample2/omr_marker.jpg
new file mode 100644
index 00000000..0929feec
Binary files /dev/null and b/src/tests/test_samples/sample2/omr_marker.jpg differ
diff --git a/src/tests/test_samples/sample2/sample.jpg b/src/tests/test_samples/sample2/sample.jpg
new file mode 100644
index 00000000..2eef7c32
Binary files /dev/null and b/src/tests/test_samples/sample2/sample.jpg differ
diff --git a/src/tests/test_template_validations.py b/src/tests/test_template_validations.py
new file mode 100644
index 00000000..bd321a72
--- /dev/null
+++ b/src/tests/test_template_validations.py
@@ -0,0 +1,159 @@
+import os
+from pathlib import Path
+
+from src.tests.test_samples.sample1.boilerplate import TEMPLATE_BOILERPLATE
+from src.tests.utils import (
+ generate_write_jsons_and_run,
+ run_entry_point,
+ setup_mocker_patches,
+)
+
+FROZEN_TIMESTAMP = "1970-01-01"
+CURRENT_DIR = Path("src/tests")
+BASE_SAMPLE_PATH = CURRENT_DIR.joinpath("test_samples", "sample1")
+BASE_SAMPLE_TEMPLATE_PATH = BASE_SAMPLE_PATH.joinpath("template.json")
+
+
+def run_sample(mocker, input_path):
+ setup_mocker_patches(mocker)
+ output_dir = os.path.join("outputs", input_path)
+ run_entry_point(input_path, output_dir)
+
+
+write_jsons_and_run = generate_write_jsons_and_run(
+ run_sample,
+ sample_path=BASE_SAMPLE_PATH,
+ template_boilerplate=TEMPLATE_BOILERPLATE,
+)
+
+
+def test_no_input_dir(mocker):
+ try:
+ run_sample(mocker, "X")
+ except Exception as e:
+ assert str(e) == "Given input directory does not exist: 'X'"
+
+
+def test_no_template(mocker):
+ if os.path.exists(BASE_SAMPLE_TEMPLATE_PATH):
+ os.remove(BASE_SAMPLE_TEMPLATE_PATH)
+ try:
+ run_sample(mocker, BASE_SAMPLE_PATH)
+ except Exception as e:
+ assert (
+ str(e)
+ == "No template file found in the directory tree of src/tests/test_samples/sample1"
+ )
+
+
+def test_empty_template(mocker):
+ def modify_template(_):
+ return {}
+
+ exception = write_jsons_and_run(mocker, modify_template=modify_template)
+ assert (
+ str(exception)
+ == f"Provided Template JSON is Invalid: '{BASE_SAMPLE_TEMPLATE_PATH}'"
+ )
+
+
+def test_invalid_field_type(mocker):
+ def modify_template(template):
+ template["fieldBlocks"]["MCQ_Block_1"]["fieldType"] = "X"
+
+ exception = write_jsons_and_run(mocker, modify_template=modify_template)
+ assert (
+ str(exception)
+ == f"Provided Template JSON is Invalid: '{BASE_SAMPLE_TEMPLATE_PATH}'"
+ )
+
+
+def test_overflow_labels(mocker):
+ def modify_template(template):
+ template["fieldBlocks"]["MCQ_Block_1"]["fieldLabels"] = ["q1..100"]
+
+ exception = write_jsons_and_run(mocker, modify_template=modify_template)
+ assert (
+ str(exception)
+ == "Overflowing field block 'MCQ_Block_1' with origin [65, 60] and dimensions [189, 5173] in template with dimensions [300, 400]"
+ )
+
+
+def test_overflow_safe_dimensions(mocker):
+ def modify_template(template):
+ template["pageDimensions"] = [255, 400]
+
+ exception = write_jsons_and_run(mocker, modify_template=modify_template)
+ assert str(exception) == "No Error"
+
+
+def test_field_strings_overlap(mocker):
+ def modify_template(template):
+ template["fieldBlocks"] = {
+ **template["fieldBlocks"],
+ "New_Block": {
+ **template["fieldBlocks"]["MCQ_Block_1"],
+ "fieldLabels": ["q5"],
+ },
+ }
+
+ exception = write_jsons_and_run(mocker, modify_template=modify_template)
+ assert str(exception) == (
+ "The field strings for field block New_Block overlap with other existing fields"
+ )
+
+
+def test_custom_label_strings_overlap_single(mocker):
+ def modify_template(template):
+ template["customLabels"] = {
+ "label1": ["q1..2", "q2..3"],
+ }
+
+ exception = write_jsons_and_run(mocker, modify_template=modify_template)
+ assert (
+ str(exception)
+ == "Given field string 'q2..3' has overlapping field(s) with other fields in 'Custom Label: label1': ['q1..2', 'q2..3']"
+ )
+
+
+def test_custom_label_strings_overlap_multiple(mocker):
+ def modify_template(template):
+ template["customLabels"] = {
+ "label1": ["q1..2"],
+ "label2": ["q2..3"],
+ }
+
+ exception = write_jsons_and_run(mocker, modify_template=modify_template)
+ assert (
+ str(exception)
+ == "The field strings for custom label 'label2' overlap with other existing custom labels"
+ )
+
+
+def test_missing_field_block_labels(mocker):
+ def modify_template(template):
+ template["customLabels"] = {"Combined": ["qX", "qY"]}
+
+ exception = write_jsons_and_run(mocker, modify_template=modify_template)
+ assert (
+ str(exception)
+ == "Missing field block label(s) in the given template for ['qX', 'qY'] from 'Combined'"
+ )
+
+
+def test_missing_output_columns(mocker):
+ def modify_template(template):
+ template["outputColumns"] = ["qX", "q1..5"]
+
+ exception = write_jsons_and_run(mocker, modify_template=modify_template)
+ assert str(exception) == (
+ "Some columns are missing in the field blocks for the given output columns"
+ )
+
+
+def test_safe_missing_label_columns(mocker):
+ def modify_template(template):
+ template["outputColumns"] = ["q1..4"]
+
+ exception = write_jsons_and_run(mocker, modify_template=modify_template)
+ assert str(exception) == "No Error"
diff --git a/src/tests/utils.py b/src/tests/utils.py
new file mode 100644
index 00000000..f8e5bfab
--- /dev/null
+++ b/src/tests/utils.py
@@ -0,0 +1,97 @@
+import json
+import os
+from copy import deepcopy
+
+from freezegun import freeze_time
+
+from main import entry_point_for_args
+
+FROZEN_TIMESTAMP = "1970-01-01"
+
+
+def setup_mocker_patches(mocker):
+ mock_imshow = mocker.patch("cv2.imshow")
+ mock_imshow.return_value = True
+
+ mock_destroy_all_windows = mocker.patch("cv2.destroyAllWindows")
+ mock_destroy_all_windows.return_value = True
+
+ mock_wait_key = mocker.patch("cv2.waitKey")
+ mock_wait_key.return_value = ord("q")
+
+
+def run_entry_point(input_path, output_dir):
+ args = {
+ "autoAlign": False,
+ "debug": False,
+ "input_paths": [input_path],
+ "output_dir": output_dir,
+ "setLayout": False,
+ "silent": True,
+ }
+ with freeze_time(FROZEN_TIMESTAMP):
+ entry_point_for_args(args)
+
+
+def write_modified(modify_content, boilerplate, sample_json_path):
+ if boilerplate is None:
+ return
+
+ content = deepcopy(boilerplate)
+
+ if modify_content is not None:
+ returned_value = modify_content(content)
+ if returned_value is not None:
+ content = returned_value
+
+ with open(sample_json_path, "w") as f:
+ json.dump(content, f)
+
+
+def remove_file(path):
+ if os.path.exists(path):
+ os.remove(path)
+
+
+def generate_write_jsons_and_run(
+ run_sample,
+ sample_path,
+ template_boilerplate=None,
+ config_boilerplate=None,
+ evaluation_boilerplate=None,
+):
+ if (template_boilerplate or config_boilerplate or evaluation_boilerplate) is None:
+ raise Exception(
+ f"No boilerplates found. Provide atleast one boilerplate to write json."
+ )
+
+ def write_jsons_and_run(
+ mocker,
+ modify_template=None,
+ modify_config=None,
+ modify_evaluation=None,
+ ):
+ sample_template_path, sample_config_path, sample_evaluation_path = (
+ sample_path.joinpath("template.json"),
+ sample_path.joinpath("config.json"),
+ sample_path.joinpath("evaluation.json"),
+ )
+ write_modified(modify_template, template_boilerplate, sample_template_path)
+ write_modified(modify_config, config_boilerplate, sample_config_path)
+ write_modified(
+ modify_evaluation, evaluation_boilerplate, sample_evaluation_path
+ )
+
+ exception = "No Error"
+ try:
+ run_sample(mocker, sample_path)
+ except Exception as e:
+ exception = e
+
+ remove_file(sample_template_path)
+ remove_file(sample_config_path)
+ remove_file(sample_evaluation_path)
+
+ return exception
+
+ return write_jsons_and_run
diff --git a/src/utils/__init__.py b/src/utils/__init__.py
new file mode 100644
index 00000000..e69de29b
diff --git a/src/utils/file.py b/src/utils/file.py
new file mode 100644
index 00000000..08536ff1
--- /dev/null
+++ b/src/utils/file.py
@@ -0,0 +1,95 @@
+import argparse
+import json
+import os
+from csv import QUOTE_NONNUMERIC
+from time import localtime, strftime
+
+import pandas as pd
+
+from src.logger import logger
+
+
+def load_json(path, **rest):
+ try:
+ with open(path, "r") as f:
+ loaded = json.load(f, **rest)
+ except json.decoder.JSONDecodeError as error:
+ logger.critical(f"Error when loading json file at: '{path}'\n{error}")
+ exit(1)
+ return loaded
+
+
+class Paths:
+ def __init__(self, output_dir):
+ self.output_dir = output_dir
+ self.save_marked_dir = output_dir.joinpath("CheckedOMRs")
+ self.results_dir = output_dir.joinpath("Results")
+ self.manual_dir = output_dir.joinpath("Manual")
+ self.evaluation_dir = output_dir.joinpath("Evaluation")
+ self.errors_dir = self.manual_dir.joinpath("ErrorFiles")
+ self.multi_marked_dir = self.manual_dir.joinpath("MultiMarkedFiles")
+
+
+def setup_dirs_for_paths(paths):
+ logger.info("Checking Directories...")
+ for save_output_dir in [paths.save_marked_dir]:
+ if not os.path.exists(save_output_dir):
+ logger.info(f"Created : {save_output_dir}")
+ os.makedirs(save_output_dir)
+ os.mkdir(save_output_dir.joinpath("stack"))
+ os.mkdir(save_output_dir.joinpath("_MULTI_"))
+ os.mkdir(save_output_dir.joinpath("_MULTI_", "stack"))
+
+ for save_output_dir in [paths.manual_dir, paths.results_dir, paths.evaluation_dir]:
+ if not os.path.exists(save_output_dir):
+ logger.info(f"Created : {save_output_dir}")
+ os.makedirs(save_output_dir)
+
+ for save_output_dir in [paths.multi_marked_dir, paths.errors_dir]:
+ if not os.path.exists(save_output_dir):
+ logger.info(f"Created : {save_output_dir}")
+ os.makedirs(save_output_dir)
+
+
+def setup_outputs_for_template(paths, template):
+ # TODO: consider moving this into a class instance
+ ns = argparse.Namespace()
+ logger.info("Checking Files...")
+
+ # Include current output paths
+ ns.paths = paths
+
+ ns.empty_resp = [""] * len(template.output_columns)
+ ns.sheetCols = [
+ "file_id",
+ "input_path",
+ "output_path",
+ "score",
+ ] + template.output_columns
+ ns.OUTPUT_SET = []
+ ns.files_obj = {}
+ TIME_NOW_HRS = strftime("%I%p", localtime())
+ ns.filesMap = {
+ "Results": os.path.join(paths.results_dir, f"Results_{TIME_NOW_HRS}.csv"),
+ "MultiMarked": os.path.join(paths.manual_dir, "MultiMarkedFiles.csv"),
+ "Errors": os.path.join(paths.manual_dir, "ErrorFiles.csv"),
+ }
+
+ for file_key, file_name in ns.filesMap.items():
+ if not os.path.exists(file_name):
+ logger.info(f"Created new file: '{file_name}'")
+ # moved handling of files to pandas csv writer
+ ns.files_obj[file_key] = file_name
+ # Create Header Columns
+ pd.DataFrame([ns.sheetCols], dtype=str).to_csv(
+ ns.files_obj[file_key],
+ mode="a",
+ quoting=QUOTE_NONNUMERIC,
+ header=False,
+ index=False,
+ )
+ else:
+ logger.info(f"Present : appending to '{file_name}'")
+ ns.files_obj[file_key] = open(file_name, "a")
+
+ return ns
diff --git a/src/utils/image.py b/src/utils/image.py
new file mode 100644
index 00000000..bba6f6d1
--- /dev/null
+++ b/src/utils/image.py
@@ -0,0 +1,155 @@
+"""
+
+ OMRChecker
+
+ Author: Udayraj Deshmukh
+ Github: https://github.com/Udayraj123
+
+"""
+import cv2
+import matplotlib.pyplot as plt
+import numpy as np
+
+from src.logger import logger
+
+plt.rcParams["figure.figsize"] = (10.0, 8.0)
+CLAHE_HELPER = cv2.createCLAHE(clipLimit=5.0, tileGridSize=(8, 8))
+
+
+class ImageUtils:
+ """A Static-only Class to hold common image processing utilities & wrappers over OpenCV functions"""
+
+ @staticmethod
+ def save_img(path, final_marked):
+ logger.info(f"Saving Image to '{path}'")
+ cv2.imwrite(path, final_marked)
+
+ @staticmethod
+ def resize_util(img, u_width, u_height=None):
+ if u_height is None:
+ h, w = img.shape[:2]
+ u_height = int(h * u_width / w)
+ return cv2.resize(img, (int(u_width), int(u_height)))
+
+ @staticmethod
+ def resize_util_h(img, u_height, u_width=None):
+ if u_width is None:
+ h, w = img.shape[:2]
+ u_width = int(w * u_height / h)
+ return cv2.resize(img, (int(u_width), int(u_height)))
+
+ @staticmethod
+ def grab_contours(cnts):
+ # source: imutils package
+
+ # if the length the contours tuple returned by cv2.findContours
+ # is '2' then we are using either OpenCV v2.4, v4-beta, or
+ # v4-official
+ if len(cnts) == 2:
+ cnts = cnts[0]
+
+ # if the length of the contours tuple is '3' then we are using
+ # either OpenCV v3, v4-pre, or v4-alpha
+ elif len(cnts) == 3:
+ cnts = cnts[1]
+
+ # otherwise OpenCV has changed their cv2.findContours return
+ # signature yet again and I have no idea WTH is going on
+ else:
+ raise Exception(
+ (
+ "Contours tuple must have length 2 or 3, "
+ "otherwise OpenCV changed their cv2.findContours return "
+ "signature yet again. Refer to OpenCV's documentation "
+ "in that case"
+ )
+ )
+
+ # return the actual contours array
+ return cnts
+
+ @staticmethod
+ def normalize_util(img, alpha=0, beta=255):
+ return cv2.normalize(img, alpha, beta, norm_type=cv2.NORM_MINMAX)
+
+ @staticmethod
+ def auto_canny(image, sigma=0.93):
+ # compute the median of the single channel pixel intensities
+ v = np.median(image)
+
+ # apply automatic Canny edge detection using the computed median
+ lower = int(max(0, (1.0 - sigma) * v))
+ upper = int(min(255, (1.0 + sigma) * v))
+ edged = cv2.Canny(image, lower, upper)
+
+ # return the edged image
+ return edged
+
+ @staticmethod
+ def adjust_gamma(image, gamma=1.0):
+ # build a lookup table mapping the pixel values [0, 255] to
+ # their adjusted gamma values
+ inv_gamma = 1.0 / gamma
+ table = np.array(
+ [((i / 255.0) ** inv_gamma) * 255 for i in np.arange(0, 256)]
+ ).astype("uint8")
+
+ # apply gamma correction using the lookup table
+ return cv2.LUT(image, table)
+
+ @staticmethod
+ def four_point_transform(image, pts):
+ # obtain a consistent order of the points and unpack them
+ # individually
+ rect = ImageUtils.order_points(pts)
+ (tl, tr, br, bl) = rect
+
+ # compute the width of the new image, which will be the
+ width_a = np.sqrt(((br[0] - bl[0]) ** 2) + ((br[1] - bl[1]) ** 2))
+ width_b = np.sqrt(((tr[0] - tl[0]) ** 2) + ((tr[1] - tl[1]) ** 2))
+
+ max_width = max(int(width_a), int(width_b))
+ # max_width = max(int(np.linalg.norm(br-bl)), int(np.linalg.norm(tr-tl)))
+
+ # compute the height of the new image, which will be the
+ height_a = np.sqrt(((tr[0] - br[0]) ** 2) + ((tr[1] - br[1]) ** 2))
+ height_b = np.sqrt(((tl[0] - bl[0]) ** 2) + ((tl[1] - bl[1]) ** 2))
+ max_height = max(int(height_a), int(height_b))
+ # max_height = max(int(np.linalg.norm(tr-br)), int(np.linalg.norm(tl-br)))
+
+ # now that we have the dimensions of the new image, construct
+ # the set of destination points to obtain a "birds eye view",
+ # (i.e. top-down view) of the image, again specifying points
+ # in the top-left, top-right, bottom-right, and bottom-left
+ # order
+ dst = np.array(
+ [
+ [0, 0],
+ [max_width - 1, 0],
+ [max_width - 1, max_height - 1],
+ [0, max_height - 1],
+ ],
+ dtype="float32",
+ )
+
+ transform_matrix = cv2.getPerspectiveTransform(rect, dst)
+ warped = cv2.warpPerspective(image, transform_matrix, (max_width, max_height))
+
+ # return the warped image
+ return warped
+
+ @staticmethod
+ def order_points(pts):
+ rect = np.zeros((4, 2), dtype="float32")
+
+ # the top-left point will have the smallest sum, whereas
+ # the bottom-right point will have the largest sum
+ s = pts.sum(axis=1)
+ rect[0] = pts[np.argmin(s)]
+ rect[2] = pts[np.argmax(s)]
+ diff = np.diff(pts, axis=1)
+ rect[1] = pts[np.argmin(diff)]
+ rect[3] = pts[np.argmax(diff)]
+
+ # return the ordered coordinates
+ return rect
diff --git a/src/utils/interaction.py b/src/utils/interaction.py
new file mode 100644
index 00000000..aba5e754
--- /dev/null
+++ b/src/utils/interaction.py
@@ -0,0 +1,107 @@
+from dataclasses import dataclass
+
+import cv2
+from screeninfo import get_monitors
+
+from src.logger import logger
+from src.utils.image import ImageUtils
+
+monitor_window = get_monitors()[0]
+
+
+@dataclass
+class ImageMetrics:
+ # TODO: Move TEXT_SIZE, etc here and find a better class name
+ window_width, window_height = monitor_window.width, monitor_window.height
+ # for positioning image windows
+ window_x, window_y = 0, 0
+ reset_pos = [0, 0]
+
+
+class InteractionUtils:
+ """Perform primary functions such as displaying images and reading responses"""
+
+ image_metrics = ImageMetrics()
+
+ @staticmethod
+ def show(name, origin, pause=1, resize=False, reset_pos=None, config=None):
+ image_metrics = InteractionUtils.image_metrics
+ if origin is None:
+ logger.info(f"'{name}' - NoneType image to show!")
+ if pause:
+ cv2.destroyAllWindows()
+ return
+ if resize:
+ if not config:
+ raise Exception("config not provided for resizing the image to show")
+ img = ImageUtils.resize_util(origin, config.dimensions.display_width)
+ else:
+ img = origin
+
+ if not is_window_available(name):
+ cv2.namedWindow(name)
+
+ cv2.imshow(name, img)
+
+ if reset_pos:
+ image_metrics.window_x = reset_pos[0]
+ image_metrics.window_y = reset_pos[1]
+
+ cv2.moveWindow(
+ name,
+ image_metrics.window_x,
+ image_metrics.window_y,
+ )
+
+ h, w = img.shape[:2]
+
+ # Set next window position
+ margin = 25
+ w += margin
+ h += margin
+
+ w, h = w // 2, h // 2
+ if image_metrics.window_x + w > image_metrics.window_width:
+ image_metrics.window_x = 0
+ if image_metrics.window_y + h > image_metrics.window_height:
+ image_metrics.window_y = 0
+ else:
+ image_metrics.window_y += h
+ else:
+ image_metrics.window_x += w
+
+ if pause:
+ logger.info(
+ f"Showing '{name}'\n\t Press Q on image to continue. Press Ctrl + C in terminal to exit"
+ )
+
+ wait_q()
+ InteractionUtils.image_metrics.window_x = 0
+ InteractionUtils.image_metrics.window_y = 0
+
+
+@dataclass
+class Stats:
+ # TODO Fill these for stats
+ # Move qbox_vals here?
+ # badThresholds = []
+ # veryBadPoints = []
+ files_moved = 0
+ files_not_moved = 0
+
+
+def wait_q():
+ esc_key = 27
+ while cv2.waitKey(1) & 0xFF not in [ord("q"), esc_key]:
+ pass
+ cv2.destroyAllWindows()
+
+
+def is_window_available(name: str) -> bool:
+ """Checks if a window is available"""
+ try:
+ cv2.getWindowProperty(name, cv2.WND_PROP_VISIBLE)
+ return True
+ except Exception as e:
+ print(e)
+ return False
diff --git a/src/utils/parsing.py b/src/utils/parsing.py
new file mode 100644
index 00000000..cb6901c6
--- /dev/null
+++ b/src/utils/parsing.py
@@ -0,0 +1,113 @@
+import re
+from copy import deepcopy
+from fractions import Fraction
+
+from deepmerge import Merger
+from dotmap import DotMap
+
+from src.constants import FIELD_LABEL_NUMBER_REGEX
+from src.defaults import CONFIG_DEFAULTS, TEMPLATE_DEFAULTS
+from src.schemas.constants import FIELD_STRING_REGEX_GROUPS
+from src.utils.file import load_json
+from src.utils.validations import (
+ validate_config_json,
+ validate_evaluation_json,
+ validate_template_json,
+)
+
+OVERRIDE_MERGER = Merger(
+ # pass in a list of tuples,with the
+ # strategies you are looking to apply
+ # to each type.
+ [
+ # (list, ["prepend"]),
+ (dict, ["merge"])
+ ],
+ # next, choose the fallback strategies,
+ # applied to all other types:
+ ["override"],
+ # finally, choose the strategies in
+ # the case where the types conflict:
+ ["override"],
+)
+
+
+def get_concatenated_response(omr_response, template):
+ # Multi-column/multi-row questions which need to be concatenated
+ concatenated_response = {}
+ for field_label, concatenate_keys in template.custom_labels.items():
+ custom_label = "".join([omr_response[k] for k in concatenate_keys])
+ concatenated_response[field_label] = custom_label
+
+ for field_label in template.non_custom_labels:
+ concatenated_response[field_label] = omr_response[field_label]
+
+ return concatenated_response
+
+
+def open_config_with_defaults(config_path):
+ user_tuning_config = load_json(config_path)
+ user_tuning_config = OVERRIDE_MERGER.merge(
+ deepcopy(CONFIG_DEFAULTS), user_tuning_config
+ )
+ validate_config_json(user_tuning_config, config_path)
+ # https://github.com/drgrib/dotmap/issues/74
+ return DotMap(user_tuning_config, _dynamic=False)
+
+
+def open_template_with_defaults(template_path):
+ user_template = load_json(template_path)
+ user_template = OVERRIDE_MERGER.merge(deepcopy(TEMPLATE_DEFAULTS), user_template)
+ validate_template_json(user_template, template_path)
+ return user_template
+
+
+def open_evaluation_with_validation(evaluation_path):
+ user_evaluation_config = load_json(evaluation_path)
+ validate_evaluation_json(user_evaluation_config, evaluation_path)
+ return user_evaluation_config
+
+
+def parse_fields(key, fields):
+ parsed_fields = []
+ fields_set = set()
+ for field_string in fields:
+ fields_array = parse_field_string(field_string)
+ current_set = set(fields_array)
+ if not fields_set.isdisjoint(current_set):
+ raise Exception(
+ f"Given field string '{field_string}' has overlapping field(s) with other fields in '{key}': {fields}"
+ )
+ fields_set.update(current_set)
+ parsed_fields.extend(fields_array)
+ return parsed_fields
+
+
+def parse_field_string(field_string):
+ if "." in field_string:
+ field_prefix, start, end = re.findall(FIELD_STRING_REGEX_GROUPS, field_string)[
+ 0
+ ]
+ start, end = int(start), int(end)
+ if start >= end:
+ raise Exception(
+ f"Invalid range in fields string: '{field_string}', start: {start} is not less than end: {end}"
+ )
+ return [
+ f"{field_prefix}{field_number}" for field_number in range(start, end + 1)
+ ]
+ else:
+ return [field_string]
+
+
+def custom_sort_output_columns(field_label):
+ label_prefix, label_suffix = re.findall(FIELD_LABEL_NUMBER_REGEX, field_label)[0]
+ return [label_prefix, int(label_suffix) if len(label_suffix) > 0 else 0]
+
+
+def parse_float_or_fraction(result):
+ if type(result) == str and "/" in result:
+ result = float(Fraction(result))
+ else:
+ result = float(result)
+ return result
diff --git a/src/utils/validations.py b/src/utils/validations.py
new file mode 100644
index 00000000..0c2ec839
--- /dev/null
+++ b/src/utils/validations.py
@@ -0,0 +1,115 @@
+"""
+
+ OMRChecker
+
+ Author: Udayraj Deshmukh
+ Github: https://github.com/Udayraj123
+
+"""
+import re
+
+import jsonschema
+from jsonschema import validate
+from rich.table import Table
+
+from src.logger import console, logger
+from src.schemas import SCHEMA_JSONS, SCHEMA_VALIDATORS
+
+
+def validate_evaluation_json(json_data, evaluation_path):
+ logger.info(f"Loading evaluation.json: {evaluation_path}")
+ try:
+ validate(instance=json_data, schema=SCHEMA_JSONS["evaluation"])
+ except jsonschema.exceptions.ValidationError as _err: # NOQA
+ table = Table(show_lines=True)
+ table.add_column("Key", style="cyan", no_wrap=True)
+ table.add_column("Error", style="magenta")
+
+ errors = sorted(
+ SCHEMA_VALIDATORS["evaluation"].iter_errors(json_data),
+ key=lambda e: e.path,
+ )
+ for error in errors:
+ key, validator, msg = parse_validation_error(error)
+ if validator == "required":
+ requiredProperty = re.findall(r"'(.*?)'", msg)[0]
+ table.add_row(
+ f"{key}.{requiredProperty}",
+ msg + ". Make sure the spelling of the key is correct",
+ )
+ else:
+ table.add_row(key, msg)
+ console.print(table, justify="center")
+ raise Exception(
+ f"Provided Evaluation JSON is Invalid: '{evaluation_path}'"
+ ) from None
+
+
+def validate_template_json(json_data, template_path):
+ logger.info(f"Loading template.json: {template_path}")
+ try:
+ validate(instance=json_data, schema=SCHEMA_JSONS["template"])
+ except jsonschema.exceptions.ValidationError as _err: # NOQA
+ table = Table(show_lines=True)
+ table.add_column("Key", style="cyan", no_wrap=True)
+ table.add_column("Error", style="magenta")
+
+ errors = sorted(
+ SCHEMA_VALIDATORS["template"].iter_errors(json_data),
+ key=lambda e: e.path,
+ )
+ for error in errors:
+ key, validator, msg = parse_validation_error(error)
+
+ # Print preProcessor name in case of options error
+ if key == "preProcessors":
+ preProcessorName = json_data["preProcessors"][error.path[1]]["name"]
+ preProcessorKey = error.path[2]
+ table.add_row(f"{key}.{preProcessorName}.{preProcessorKey}", msg)
+ elif validator == "required":
+ requiredProperty = re.findall(r"'(.*?)'", msg)[0]
+ table.add_row(
+ f"{key}.{requiredProperty}",
+ f"{msg}. Check for spelling errors and make sure it is in camelCase",
+ )
+ else:
+ table.add_row(key, msg)
+ console.print(table, justify="center")
+ raise Exception(
+ f"Provided Template JSON is Invalid: '{template_path}'"
+ ) from None
+
+
+def validate_config_json(json_data, config_path):
+ logger.info(f"Loading config.json: {config_path}")
+ try:
+ validate(instance=json_data, schema=SCHEMA_JSONS["config"])
+ except jsonschema.exceptions.ValidationError as _err: # NOQA
+ table = Table(show_lines=True)
+ table.add_column("Key", style="cyan", no_wrap=True)
+ table.add_column("Error", style="magenta")
+ errors = sorted(
+ SCHEMA_VALIDATORS["config"].iter_errors(json_data),
+ key=lambda e: e.path,
+ )
+ for error in errors:
+ key, validator, msg = parse_validation_error(error)
+
+ if validator == "required":
+ requiredProperty = re.findall(r"'(.*?)'", msg)[0]
+ table.add_row(
+ f"{key}.{requiredProperty}",
+ f"{msg}. Check for spelling errors and make sure it is in camelCase",
+ )
+ else:
+ table.add_row(key, msg)
+ console.print(table, justify="center")
+ raise Exception(f"Provided config JSON is Invalid: '{config_path}'") from None
+
+
+def parse_validation_error(error):
+ return (
+ (error.path[0] if len(error.path) > 0 else "$root"),
+ error.validator,
+ error.message,
+ )
diff --git a/template.py b/template.py
deleted file mode 100644
index cdfdb276..00000000
--- a/template.py
+++ /dev/null
@@ -1,355 +0,0 @@
-"""
-
-Designed and Developed by-
-Udayraj Deshmukh
-https://github.com/Udayraj123
-
-"""
-import cv2
-import os
-import json
-import numpy as np
-from globals import *
-from utils import *
-
-### Coordinates Part ###
-
-
-class Pt():
- """
- Container for a Point Box on the OMR
- """
- """
- qNo is the point's property- question to which this point belongs to
- It can be used as a roll number column as well. (eg roll1)
- It can also correspond to a single digit of integer type Q (eg q5d1)
- """
-
- def __init__(self, pt, qNo, qType, val):
- self.x = round(pt[0])
- self.y = round(pt[1])
- self.qNo = qNo
- self.qType = qType
- self.val = val
-
-
-class QBlock():
- def __init__(self, dims, key, orig, traverse_pts):
- # dims = (width, height)
- self.dims = tuple(round(x) for x in dims)
- self.key = key
- self.orig = orig
- self.traverse_pts = traverse_pts
- # will be set when using
- self.shift = 0
-
-
-qtype_data = {
- 'QTYPE_MED': {
- 'vals': ['E', 'H'],
- 'orient': 'V'
- },
- 'QTYPE_ROLL': {
- 'vals': range(10),
- 'orient': 'V'
- },
- 'QTYPE_INT': {
- 'vals': range(10),
- 'orient': 'V'
- },
- 'QTYPE_MCQ4': {
- 'vals': ['A', 'B', 'C', 'D'],
- 'orient': 'H'
- },
- 'QTYPE_MCQ5': {
- 'vals': ['A', 'B', 'C', 'D', 'E'],
- 'orient': 'H'
- },
- # Add custom question types here-
- # ,
- # 'QTYPE_MCQ_COL_5A':{'vals' : ['A']*5, 'orient':'V'},
- # 'QTYPE_MCQ_COL_5B':{'vals' : ['B']*5, 'orient':'V'},
- # 'QTYPE_MCQ_COL_5C':{'vals' : ['C']*5, 'orient':'V'},
- # 'QTYPE_MCQ_COL_5D':{'vals' : ['D']*5, 'orient':'V'},
- # 'QTYPE_MCQ_COL_4A':{'vals' : ['A']*4, 'orient':'V'},
- # 'QTYPE_MCQ_COL_4B':{'vals' : ['B']*4, 'orient':'V'},
- # 'QTYPE_MCQ_COL_4C':{'vals' : ['C']*4, 'orient':'V'},
- # 'QTYPE_MCQ_COL_4D':{'vals' : ['D']*4, 'orient':'V'},
-}
-
-
-class Template():
- def __init__(self, path):
- with open(path, "r") as f:
- json_obj = json.load(f)
- self.path = path
- self.QBlocks = []
- # throw exception on key not exist
- self.dims = json_obj["Dimensions"]
- self.bubbleDims = json_obj["BubbleDimensions"]
- self.concats = json_obj["Concatenations"]
- self.singles = json_obj["Singles"]
-
- # Add new qTypes from template
- if "qTypes" in json_obj:
- qtype_data.update(json_obj["qTypes"])
-
- # process local options
- self.options = json_obj.get("Options", {})
-
- self.marker = None
- self.marker_path = None
- # process markers
- if "Marker" in self.options:
- markerOps = self.options["Marker"]
- self.marker_path = os.path.join(
- os.path.dirname(path), markerOps.get(
- "RelativePath", MARKER_FILE))
- if(not os.path.exists(self.marker_path)):
- print(
- "Error: Marker not found at path provided in template:",
- self.marker_path)
- exit(31)
-
- marker = cv2.imread(self.marker_path, cv2.IMREAD_GRAYSCALE)
- if("SheetToMarkerWidthRatio" in markerOps):
- marker = resize_util(marker, uniform_width /
- int(markerOps["SheetToMarkerWidthRatio"]))
- marker = cv2.GaussianBlur(marker, (5, 5), 0)
- marker = cv2.normalize(
- marker,
- None,
- alpha=0,
- beta=255,
- norm_type=cv2.NORM_MINMAX)
- # marker_eroded_sub = marker-cv2.erode(marker,None)
- self.marker = marker - \
- cv2.erode(marker, kernel=np.ones((5, 5)), iterations=5)
-
- # Allow template to override globals
- # TODO: This is a hack as there should no be any global configuration
- # All template configuration should be local. Global config should
- # be via command args.
- self.globals = json_obj.get("Globals")
- self.update_globals()
-
- # Add QBlocks
- for name, block in json_obj["QBlocks"].items():
- self.addQBlocks(name, block)
-
- def update_globals(self):
- if self.globals:
- globals().update(self.globals)
-
- # Expects bubbleDims to be set already
- def addQBlocks(self, key, rect):
- assert(self.bubbleDims != [-1, -1])
- # For qType defined in QBlocks
- if 'qType' in rect:
- rect.update(**qtype_data[rect['qType']])
- else:
- rect['qType'] = {'vals': rect['vals'], 'orient': rect['orient']}
- # keyword arg unpacking followed by named args
- self.QBlocks += genGrid(self.bubbleDims, key, **rect)
- # self.QBlocks.append(QBlock(rect.orig, calcQBlockDims(rect), maketemplate(rect)))
-
-
-def genQBlock(
- bubbleDims,
- QBlockDims,
- key,
- orig,
- qNos,
- gaps,
- vals,
- qType,
- orient,
- col_orient):
- """
- Input:
- orig - start point
- qNos - a tuple of qNos
- gaps - (gapX,gapY) are the gaps between rows and cols in a block
- vals - a 1D array of values of each alternative for a question
-
- Output:
- // Returns set of coordinates of a rectangular grid of points
- Returns a QBlock containing array of Qs and some metadata?!
-
- Ref:
- 1 2 3 4
- 1 2 3 4
- 1 2 3 4
-
- (q1, q2, q3)
-
- 00
- 11
- 22
- 33
- 44
-
- (q1.1,q1.2)
-
- """
- H, V = (0, 1) if(orient == 'H') else (1, 0)
- # orig[0] += np.random.randint(-6,6)*2 # test random shift
- traverse_pts = []
- o = [float(i) for i in orig]
-
- if(col_orient == orient):
- for q in range(len(qNos)):
- pt = o.copy()
- pts = []
- for v in range(len(vals)):
- pts.append(Pt(pt.copy(), qNos[q], qType, vals[v]))
- pt[H] += gaps[H]
- # For diagonalal endpoint of QBlock
- pt[H] += bubbleDims[H] - gaps[H]
- pt[V] += bubbleDims[V]
- # TODO- make a mini object for this
- traverse_pts.append(([o.copy(), pt.copy()], pts))
- o[V] += gaps[V]
- else:
- for v in range(len(vals)):
- pt = o.copy()
- pts = []
- for q in range(len(qNos)):
- pts.append(Pt(pt.copy(), qNos[q], qType, vals[v]))
- pt[V] += gaps[V]
- # For diagonalal endpoint of QBlock
- pt[V] += bubbleDims[V] - gaps[V]
- pt[H] += bubbleDims[H]
- # TODO- make a mini object for this
- traverse_pts.append(([o.copy(), pt.copy()], pts))
- o[H] += gaps[H]
- # Pass first three args as is. only append 'traverse_pts'
- return QBlock(QBlockDims, key, orig, traverse_pts)
-
-
-def genGrid(
- bubbleDims,
- key,
- qType,
- orig,
- bigGaps,
- gaps,
- qNos,
- vals,
- orient='V',
- col_orient='V'):
- """
- Input(Directly passable from JSON parameters):
- bubbleDims - dimesions of single QBox
- orig- start point
- qNos - an array of qNos tuples(see below) that align with dimension of the big grid (gridDims extracted from here)
- bigGaps - (bigGapX,bigGapY) are the gaps between blocks
- gaps - (gapX,gapY) are the gaps between rows and cols in a block
- vals - a 1D array of values of each alternative for a question
- orient - The way of arranging the vals (vertical or horizontal)
-
- Output:
- // Returns an array of Q objects (having their points) arranged in a rectangular grid
- Returns grid of QBlock objects
-
- 00 00 00 00
- Q1 1 2 3 4 1 2 3 4 11 11 11 11
- Q2 1 2 3 4 1 2 3 4 22 22 22 22 1234567
- Q3 1 2 3 4 1 2 3 4 33 33 33 33 1234567
- 44 44 44 44
- , 55 55 55 55 , 1234567 and many more possibilities!
- Q7 1 2 3 4 1 2 3 4 66 66 66 66 1234567
- Q8 1 2 3 4 1 2 3 4 77 77 77 77
- Q9 1 2 3 4 1 2 3 4 88 88 88 88
- 99 99 99 99
-
-TODO: Update this part, add more examples like-
- Q1 1 2 3 4
-
- Q2 1 2 3 4
- Q3 1 2 3 4
-
- Q4 1 2 3 4
- Q5 1 2 3 4
-
- MCQ type (orient='H')-
- [
- [(q1,q2,q3),(q4,q5,q6)]
- [(q7,q8,q9),(q10,q11,q12)]
- ]
-
- INT type (orient='V')-
- [
- [(q1d1,q1d2),(q2d1,q2d2),(q3d1,q3d2),(q4d1,q4d2)]
- ]
-
- ROLL type-
- [
- [(roll1,roll2,roll3,...,roll10)]
- ]
-
- """
- gridData = np.array(qNos)
- # print(gridData.shape, gridData)
- if(0 and len(gridData.shape) != 3 or gridData.size == 0): # product of shape is zero
- print(
- "Error(genGrid): Invalid qNos array given:",
- gridData.shape,
- gridData)
- exit(32)
-
- orig = np.array(orig)
- numQsMax = max([max([len(qb) for qb in row]) for row in gridData])
-
- numDims = [numQsMax, len(vals)]
-
- QBlocks = []
-
- # **Simple is powerful**
- # H and V are named with respect to orient == 'H', reverse their meaning
- # when orient = 'V'
- H, V = (0, 1) if(orient == 'H') else (1, 0)
-
- # print(orig, numDims, gridData.shape, gridData)
- # orient is also the direction of making QBlocks
-
- # print(key, numDims, orig, gaps, bigGaps, origGap )
- qStart = orig.copy()
-
- origGap = [0, 0]
-
- # Usually single row
- for row in gridData:
- qStart[V] = orig[V]
-
- # Usually multiple qTuples
- for qTuple in row:
- # Update numDims and origGaps
- numDims[0] = len(qTuple)
- # bigGaps is indep of orientation
- origGap[0] = bigGaps[0] + (numDims[V] - 1) * gaps[H]
- origGap[1] = bigGaps[1] + (numDims[H] - 1) * gaps[V]
- # each qTuple will have qNos
- QBlockDims = [
- # width x height in pixels
- gaps[0] * (numDims[V] - 1) + bubbleDims[H],
- gaps[1] * (numDims[H] - 1) + bubbleDims[V]
- ]
- # WATCH FOR BLUNDER(use .copy()) - qStart was getting passed by
- # reference! (others args read-only)
- QBlocks.append(
- genQBlock(
- bubbleDims,
- QBlockDims,
- key,
- qStart.copy(),
- qTuple,
- gaps,
- vals,
- qType,
- orient,
- col_orient))
- # Goes vertically down first
- qStart[V] += origGap[V]
- qStart[H] += origGap[H]
- return QBlocks
diff --git a/utils.py b/utils.py
deleted file mode 100644
index 25f6df64..00000000
--- a/utils.py
+++ /dev/null
@@ -1,1168 +0,0 @@
-"""
-
-Designed and Developed by-
-Udayraj Deshmukh
-https://github.com/Udayraj123
-
-"""
-from time import localtime, strftime, time
-from random import randint
-from template import *
-from globals import *
-
-# In[62]:
-import re
-import os
-import sys
-import cv2
-import glob
-from imutils import grab_contours
-import numpy as np
-import pandas as pd
-import matplotlib.pyplot as plt
-plt.rcParams['figure.figsize'] = (10.0, 8.0)
-
-
-def setup_dirs(paths):
- print('\nChecking Directories...')
- for _dir in [paths.saveMarkedDir]:
- if(not os.path.exists(_dir)):
- print('Created : ' + _dir)
- os.makedirs(_dir)
- os.mkdir(_dir + '/stack')
- os.mkdir(_dir + '/_MULTI_')
- os.mkdir(_dir + '/_MULTI_' + '/stack')
- # os.mkdir(_dir+sl+'/_BADSCAN_')
- # os.mkdir(_dir+sl+'/_BADSCAN_'+'/stack')
- else:
- print('Present : ' + _dir)
-
- for _dir in [paths.manualDir, paths.resultDir]:
- if(not os.path.exists(_dir)):
- print('Created : ' + _dir)
- os.makedirs(_dir)
- else:
- print('Present : ' + _dir)
-
- for _dir in [paths.multiMarkedDir, paths.errorsDir, paths.badRollsDir]:
- if(not os.path.exists(_dir)):
- print('Created : ' + _dir)
- os.makedirs(_dir)
- else:
- print('Present : ' + _dir)
-
-
-def waitQ():
- ESC_KEY = 27
- while(cv2.waitKey(1) & 0xFF not in [ord('q'), ESC_KEY]):
- pass
- cv2.destroyAllWindows()
-
-
-def normalize_util(img, alpha=0, beta=255):
- return cv2.normalize(img, alpha, beta, norm_type=cv2.NORM_MINMAX)
-
-
-def normalize_hist(img):
- hist, bins = np.histogram(img.flatten(), 256, [0, 256])
- cdf = hist.cumsum()
- cdf_m = np.ma.masked_equal(cdf, 0)
- cdf_m = (cdf_m - cdf_m.min()) * 255 / (cdf_m.max() - cdf_m.min())
- cdf = np.ma.filled(cdf_m, 0).astype('uint8')
- return cdf[img]
-
-
-def resize_util(img, u_width, u_height=None):
- if u_height is None:
- h, w = img.shape[:2]
- u_height = int(h * u_width / w)
- return cv2.resize(img, (int(u_width), int(u_height)))
-
-
-def resize_util_h(img, u_height, u_width=None):
- if u_width is None:
- h, w = img.shape[:2]
- u_width = int(w * u_height / h)
- return cv2.resize(img, (int(u_width), int(u_height)))
-
-
-def show(name, orig, pause=1, resize=False, resetpos=None):
- global windowX, windowY, display_width
- if(type(orig) == type(None)):
- print(name, " NoneType image to show!")
- if(pause):
- cv2.destroyAllWindows()
- return
- origDim = orig.shape[:2]
- img = resize_util(orig, display_width, display_height) if resize else orig
- cv2.imshow(name, img)
- if(resetpos):
- windowX = resetpos[0]
- windowY = resetpos[1]
- cv2.moveWindow(name, windowX, windowY)
-
- h, w = img.shape[:2]
-
- # Set next window position
- margin = 25
- w += margin
- h += margin
- if(windowX + w > windowWidth):
- windowX = 0
- if(windowY + h > windowHeight):
- windowY = 0
- else:
- windowY += h
- else:
- windowX += w
-
- if(pause):
- print(
- "Showing '" +
- name +
- "'\n\tPress Q on image to continue Press Ctrl + C in terminal to exit")
- waitQ()
-
-
-def putLabel(img, label, size):
- scale = img.shape[1] / display_width
- bgVal = int(np.mean(img))
- pos = (int(scale * 80), int(scale * 30))
- clr = (255 - bgVal,) * 3
- img[(pos[1] - size * 30):(pos[1] + size * 2), :] = bgVal
- cv2.putText(img, label, pos, cv2.FONT_HERSHEY_SIMPLEX, size, clr, 3)
-
-
-def drawTemplateLayout(
- img,
- template,
- shifted=True,
- draw_qvals=False,
- border=-1):
- img = resize_util(img, template.dims[0], template.dims[1])
- final_align = img.copy()
- boxW, boxH = template.bubbleDims
- for QBlock in template.QBlocks:
- s, d = QBlock.orig, QBlock.dims
- shift = QBlock.shift
- if(shifted):
- cv2.rectangle(
- final_align,
- (s[0] + shift,
- s[1]),
- (s[0] + shift + d[0],
- s[1] + d[1]),
- CLR_BLACK,
- 3)
- else:
- cv2.rectangle(
- final_align, (s[0], s[1]), (s[0] + d[0], s[1] + d[1]), CLR_BLACK, 3)
- for qStrip, qBoxPts in QBlock.traverse_pts:
- for pt in qBoxPts:
- x, y = (pt.x + QBlock.shift, pt.y) if shifted else (pt.x, pt.y)
- cv2.rectangle(final_align,
- (int(x + boxW / 10),
- int(y + boxH / 10)),
- (int(x + boxW - boxW / 10),
- int(y + boxH - boxH / 10)),
- CLR_GRAY,
- border)
- if(draw_qvals):
- rect = [y, y + boxH, x, x + boxW]
- cv2.putText(final_align, '%d' % (cv2.mean(img[rect[0]:rect[1], rect[2]:rect[3]])[
- 0]), (rect[2] + 2, rect[0] + (boxH * 2) // 3), cv2.FONT_HERSHEY_SIMPLEX, 0.6, CLR_BLACK, 2)
- if(shifted):
- cv2.putText(final_align, 's%s' %
- (shift), tuple(s -
- [template.dims[0] //
- 20, -
- d[1] //
- 2]), cv2.FONT_HERSHEY_SIMPLEX, TEXT_SIZE, CLR_BLACK, 4)
- return final_align
-
-
-def getPlotImg():
- plt.savefig('tmp.png')
- # img = cv2.imread('tmp.png',cv2.IMREAD_COLOR)
- img = cv2.imread('tmp.png', cv2.IMREAD_GRAYSCALE)
- os.remove("tmp.png")
- # plt.cla()
- # plt.clf()
- plt.close()
- return img
-
-
-def order_points(pts):
- rect = np.zeros((4, 2), dtype="float32")
-
- # the top-left point will have the smallest sum, whereas
- # the bottom-right point will have the largest sum
- s = pts.sum(axis=1)
- rect[0] = pts[np.argmin(s)]
- rect[2] = pts[np.argmax(s)]
- diff = np.diff(pts, axis=1)
- rect[1] = pts[np.argmin(diff)]
- rect[3] = pts[np.argmax(diff)]
-
- # return the ordered coordinates
- return rect
-
-
-def four_point_transform(image, pts):
- # obtain a consistent order of the points and unpack them
- # individually
- rect = order_points(pts)
- (tl, tr, br, bl) = rect
-
- # compute the width of the new image, which will be the
- widthA = np.sqrt(((br[0] - bl[0]) ** 2) + ((br[1] - bl[1]) ** 2))
- widthB = np.sqrt(((tr[0] - tl[0]) ** 2) + ((tr[1] - tl[1]) ** 2))
-
- maxWidth = max(int(widthA), int(widthB))
- # maxWidth = max(int(np.linalg.norm(br-bl)), int(np.linalg.norm(tr-tl)))
-
- # compute the height of the new image, which will be the
- heightA = np.sqrt(((tr[0] - br[0]) ** 2) + ((tr[1] - br[1]) ** 2))
- heightB = np.sqrt(((tl[0] - bl[0]) ** 2) + ((tl[1] - bl[1]) ** 2))
- maxHeight = max(int(heightA), int(heightB))
- # maxHeight = max(int(np.linalg.norm(tr-br)), int(np.linalg.norm(tl-br)))
-
- # now that we have the dimensions of the new image, construct
- # the set of destination points to obtain a "birds eye view",
- # (i.e. top-down view) of the image, again specifying points
- # in the top-left, top-right, bottom-right, and bottom-left
- # order
- dst = np.array([
- [0, 0],
- [maxWidth - 1, 0],
- [maxWidth - 1, maxHeight - 1],
- [0, maxHeight - 1]], dtype="float32")
-
- # compute the perspective transform matrix and then apply it
- M = cv2.getPerspectiveTransform(rect, dst)
- warped = cv2.warpPerspective(image, M, (maxWidth, maxHeight))
-
- # return the warped image
- return warped
-
-
-def dist(p1, p2):
- return np.linalg.norm(np.array(p1) - np.array(p2))
-
-
-def get_reflection(pt, pt1, pt2):
- pt, pt1, pt2 = tuple(
- map(lambda x: np.array(x, dtype=float), [pt, pt1, pt2]))
- return (pt1 + pt2) - pt
-
-
-def printbuf(x):
- sys.stdout.write(str(x))
- sys.stdout.write('\r')
-
-
-def get_fourth_pt(three_pts):
- m = []
- for i in range(3):
- m.append(dist(three_pts[i], three_pts[(i + 1) % 3]))
-
- v = max(m)
- for i in range(3):
- if(m[i] != v and m[(i + 1) % 3] != v):
- refl = (i + 1) % 3
- break
- fourth_pt = get_reflection(
- three_pts[refl], three_pts[(refl + 1) % 3], three_pts[(refl + 2) % 3])
- return fourth_pt
-
-
-def angle(p1, p2, p0):
- dx1 = float(p1[0] - p0[0])
- dy1 = float(p1[1] - p0[1])
- dx2 = float(p2[0] - p0[0])
- dy2 = float(p2[1] - p0[1])
- return (dx1 * dx2 + dy1 * dy2) / \
- np.sqrt((dx1 * dx1 + dy1 * dy1) * (dx2 * dx2 + dy2 * dy2) + 1e-10)
-
-
-def checkMaxCosine(approx):
- # assumes 4 pts present
- maxCosine = 0
- minCosine = 1.5
- for i in range(2, 5):
- cosine = abs(angle(approx[i % 4], approx[i - 2], approx[i - 1]))
- maxCosine = max(cosine, maxCosine)
- minCosine = min(cosine, minCosine)
- # TODO add to plot dict
- # print(maxCosine)
- if(maxCosine >= 0.35):
- print('Quadrilateral is not a rectangle.')
- return False
- return True
-
-
-def validateRect(approx):
- # TODO: add logic from app?!
- return len(approx) == 4 and checkMaxCosine(approx.reshape(4, 2))
-
-
-def auto_canny(image, sigma=0.93):
- # compute the median of the single channel pixel intensities
- v = np.median(image)
-
- # apply automatic Canny edge detection using the computed median
- lower = int(max(0, (1.0 - sigma) * v))
- upper = int(min(255, (1.0 + sigma) * v))
- edged = cv2.Canny(image, lower, upper)
-
- # return the edged image
- return edged
-
-
-def resetSaveImg(key):
- global saveImgList
- saveImgList[key] = []
-
-
-def appendSaveImg(key, img):
- if(saveimglvl >= int(key)):
- global saveImgList
- if(key not in saveImgList):
- saveImgList[key] = []
- saveImgList[key].append(img.copy())
-
-
-def findPage(image_norm):
- # Done: find ORIGIN for the quadrants
- # Done, Auto tune! : Get canny parameters tuned
- # (https://www.pyimagesearch.com/2015/04/06/zero-parameter-automatic-canny-edge-detection-with-python-and-opencv/)
-
- image_norm = normalize_util(image_norm)
- ret, image_norm = cv2.threshold(image_norm, 200, 255, cv2.THRESH_TRUNC)
- image_norm = normalize_util(image_norm)
-
- appendSaveImg(1, image_norm)
-
- # kernel = cv2.getStructuringElement(cv2.MORPH_RECT, (3, 10))
- kernel = cv2.getStructuringElement(cv2.MORPH_RECT, (10, 10))
- """
- # Closing is reverse of Opening, Dilation followed by Erosion.
- A pixel in the original image (either 1 or 0) will be considered 1 only if all the pixels
- under the kernel is 1, otherwise it is eroded (made to zero).
- """
- # Close the small holes, i.e. Complete the edges on canny image
- closed = cv2.morphologyEx(image_norm, cv2.MORPH_CLOSE, kernel)
-
- appendSaveImg(1, closed)
-
- edge = cv2.Canny(closed, 185, 55)
-
- # findContours returns outer boundaries in CW and inner boundaries in ACW
- # order.
- cnts = grab_contours(
- cv2.findContours(
- edge,
- cv2.RETR_LIST,
- cv2.CHAIN_APPROX_SIMPLE))
- # hullify to resolve disordered curves due to noise
- cnts = [cv2.convexHull(c) for c in cnts]
- cnts = sorted(cnts, key=cv2.contourArea, reverse=True)[:5]
- sheet = []
- for c in cnts:
- if cv2.contourArea(c) < MIN_PAGE_AREA:
- continue
- peri = cv2.arcLength(c, True)
- # ez algo -
- # https://en.wikipedia.org/wiki/Ramer–Douglas–Peucker_algorithm
- approx = cv2.approxPolyDP(c, epsilon=0.025 * peri, closed=True)
- # print("Area",cv2.contourArea(c), "Peri", peri)
-
- # check its rectangle-ness:
- if(validateRect(approx)):
- sheet = np.reshape(approx, (4, -1))
- cv2.drawContours(image_norm, [approx], -1, (0, 255, 0), 2)
- cv2.drawContours(edge, [approx], -1, (255, 255, 255), 10)
- break
- # box = perspective.order_points(box)
- # sobel = cv2.addWeighted(cv2.Sobel(edge, cv2.CV_64F, 1, 0, ksize=3),0.5,cv2.Sobel(edge, cv2.CV_64F, 0, 1, ksize=3),0.5,0,edge)
- # ExcessDo : make it work on killer images
- # edge2 = auto_canny(image_norm)
- # show('Morphed Edges',np.hstack((closed,edge)),1,1)
-
- appendSaveImg(1, edge)
- return sheet
-
-
-# Resizing the marker within scaleRange at rate of descent_per_step to
-# find the best match.
-def getBestMatch(image_eroded_sub, marker):
-
- descent_per_step = (
- marker_rescale_range[1] - marker_rescale_range[0]) // marker_rescale_steps
- h, w = marker.shape[:2]
- res, best_scale = None, None
- allMaxT = 0
-
- for r0 in np.arange(
- marker_rescale_range[1], marker_rescale_range[0], -1 * descent_per_step): # reverse order
- s = float(r0 * 1 / 100)
- if(s == 0.0):
- continue
- rescaled_marker = resize_util_h(
- marker if ERODE_SUB_OFF else marker,
- u_height=int(
- h * s))
- # res is the black image with white dots
- res = cv2.matchTemplate(
- image_eroded_sub,
- rescaled_marker,
- cv2.TM_CCOEFF_NORMED)
-
- maxT = res.max()
- if(allMaxT < maxT):
- # print('Scale: '+str(s)+', Circle Match: '+str(round(maxT*100,2))+'%')
- best_scale, allMaxT = s, maxT
-
- if(allMaxT < thresholdCircle):
- print("\tWarning: Template matching too low! Should you pass --noCropping flag?")
- if(showimglvl >= 1):
- show("res", res, 1, 0)
-
- if(best_scale is None):
- print("No matchings for given scaleRange:", marker_rescale_range)
- return best_scale, allMaxT
-
-
-def adjust_gamma(image, gamma=1.0):
- # build a lookup table mapping the pixel values [0, 255] to
- # their adjusted gamma values
- invGamma = 1.0 / gamma
- table = np.array([((i / 255.0) ** invGamma) * 255
- for i in np.arange(0, 256)]).astype("uint8")
-
- # apply gamma correction using the lookup table
- return cv2.LUT(image, table)
-
-
-clahe = cv2.createCLAHE(clipLimit=5.0, tileGridSize=(8, 8))
-# TODO Fill these for stats
-thresholdCircles = []
-badThresholds = []
-veryBadPoints = []
-
-
-def getROI(image, filename, noCropping=False):
- global clahe
- for i in range(saveimglvl):
- resetSaveImg(i + 1)
-
- appendSaveImg(1, image)
- """
- TODO later Autorotate:
- - Rotate 90 : check page width:height, CW/ACW? - do CW, then pass to 180 check.
- - Rotate 180 :
- Nope, OMR specific, paper warping may be imperfect. - check markers centroid
- Nope - OCR check
- Match logo - can work, but 'lon' too big and may unnecessarily rotate? - but you know the scale
- Check roll field morphed
- """
-
- # TODO: (remove noCropping bool) Automate the case of close up scan(incorrect page)-
- # ^Note: App rejects croppeds along with others
-
- # image = resize_util(image, uniform_width, uniform_height)
-
- # Preprocessing the image
- img = image.copy()
- # TODO: need to detect if image is too blurry already! (M1: check
- # noCropping dimensions b4 resizing coz it won't be blurry otherwise _/)
- img = cv2.GaussianBlur(img, (3, 3), 0)
- image_norm = normalize_util(img)
-
- if(noCropping == False):
- # Need this resize for arbitrary high res images: before passing to
- # findPage
- if(image_norm.shape[1] > uniform_width * 2):
- image_norm = resize_util(image_norm, uniform_width * 2)
- sheet = findPage(image_norm)
- if sheet == []:
- print("\tError: Paper boundary not found! Should you pass --noCropping flag?")
- return None
- else:
- print("Found page corners: \t", sheet.tolist())
-
- # Warp layer 1
- image_norm = four_point_transform(image_norm, sheet)
-
- # Resize only after cropping the page for clarity as well as uniformity
- # for non noCropping images
- image_norm = resize_util(image_norm, uniform_width, uniform_height)
- image = resize_util(image, uniform_width, uniform_height)
- appendSaveImg(1, image_norm)
-
- # Return preprocessed image
- return image_norm
-
-
-def handle_markers(image_norm, marker, curr_filename):
-
- if ERODE_SUB_OFF:
- image_eroded_sub = normalize_util(image_norm)
- else:
- image_eroded_sub = normalize_util(image_norm
- - cv2.erode(image_norm,
- kernel=np.ones((5, 5)),
- iterations=5))
- # Quads on warped image
- quads = {}
- h1, w1 = image_eroded_sub.shape[:2]
- midh, midw = h1 // 3, w1 // 2
- origins = [[0, 0], [midw, 0], [0, midh], [midw, midh]]
- quads[0] = image_eroded_sub[0:midh, 0:midw]
- quads[1] = image_eroded_sub[0:midh, midw:w1]
- quads[2] = image_eroded_sub[midh:h1, 0:midw]
- quads[3] = image_eroded_sub[midh:h1, midw:w1]
-
- # Draw Quadlines
- image_eroded_sub[:, midw:midw + 2] = 255
- image_eroded_sub[midh:midh + 2, :] = 255
-
- best_scale, allMaxT = getBestMatch(image_eroded_sub, marker)
- if(best_scale is None):
- # TODO: Plot and see performance of marker_rescale_range
- if(showimglvl >= 1):
- show('Quads', image_eroded_sub)
- return None
-
- optimal_marker = resize_util_h(
- marker if ERODE_SUB_OFF else marker, u_height=int(
- marker.shape[0] * best_scale))
- h, w = optimal_marker.shape[:2]
- centres = []
- sumT, maxT = 0, 0
- print("Matching Marker:\t", end=" ")
- for k in range(0, 4):
- res = cv2.matchTemplate(quads[k], optimal_marker, cv2.TM_CCOEFF_NORMED)
- maxT = res.max()
- print("Q" + str(k + 1) + ": maxT", round(maxT, 3), end="\t")
- if(maxT < thresholdCircle or abs(allMaxT - maxT) >= thresholdVar):
- # Warning - code will stop in the middle. Keep Threshold low to
- # avoid.
- print(
- curr_filename,
- "\nError: No circle found in Quad",
- k + 1,
- "\n\tthresholdVar",
- thresholdVar,
- "maxT",
- maxT,
- "allMaxT",
- allMaxT,
- "Should you pass --noCropping flag?")
- if(showimglvl >= 1):
- show("no_pts_" + curr_filename, image_eroded_sub, 0)
- show("res_Q" + str(k + 1), res, 1)
- return None
-
- pt = np.argwhere(res == maxT)[0]
- pt = [pt[1], pt[0]]
- pt[0] += origins[k][0]
- pt[1] += origins[k][1]
- # print(">>",pt)
- image_norm = cv2.rectangle(image_norm, tuple(
- pt), (pt[0] + w, pt[1] + h), (150, 150, 150), 2)
- # display:
- image_eroded_sub = cv2.rectangle(
- image_eroded_sub,
- tuple(pt),
- (pt[0] + w,
- pt[1] + h),
- (50,
- 50,
- 50) if ERODE_SUB_OFF else (
- 155,
- 155,
- 155),
- 4)
- centres.append([pt[0] + w / 2, pt[1] + h / 2])
- sumT += maxT
- print("Optimal Scale:", best_scale)
- # analysis data
- thresholdCircles.append(sumT / 4)
-
- image_norm = four_point_transform(image_norm, np.array(centres))
- # appendSaveImg(1,image_eroded_sub)
- # appendSaveImg(1,image_norm)
-
- appendSaveImg(2, image_eroded_sub)
- # Debugging image -
- # res = cv2.matchTemplate(image_eroded_sub,optimal_marker,cv2.TM_CCOEFF_NORMED)
- # res[ : , midw:midw+2] = 255
- # res[ midh:midh+2, : ] = 255
- # show("Markers Matching",res)
- if(showimglvl >= 2 and showimglvl < 4):
- image_eroded_sub = resize_util_h(image_eroded_sub, image_norm.shape[0])
- image_eroded_sub[:, -5:] = 0
- h_stack = np.hstack((image_eroded_sub, image_norm))
- show("Warped: " + curr_filename, resize_util(h_stack,
- int(display_width * 1.6)), 0, 0, [0, 0])
- # iterations : Tuned to 2.
- # image_eroded_sub = image_norm - cv2.erode(image_norm, kernel=np.ones((5,5)),iterations=2)
- return image_norm
-
-
-def getGlobalThreshold(
- QVals_orig,
- plotTitle=None,
- plotShow=True,
- sortInPlot=True,
- looseness=1):
- """
- Note: Cannot assume qStrip has only-gray or only-white bg (in which case there is only one jump).
- So there will be either 1 or 2 jumps.
- 1 Jump :
- ......
- ||||||
- |||||| <-- risky THR
- |||||| <-- safe THR
- ....||||||
- ||||||||||
-
- 2 Jumps :
- ......
- |||||| <-- wrong THR
- ....||||||
- |||||||||| <-- safe THR
- ..||||||||||
- ||||||||||||
-
- The abstract "First LARGE GAP" is perfect for this.
- Current code is considering ONLY TOP 2 jumps(>= MIN_GAP) to be big, gives the smaller one
-
- """
- # Sort the Q vals
- QVals = sorted(QVals_orig)
- # Find the FIRST LARGE GAP and set it as threshold:
- ls = (looseness + 1) // 2
- l = len(QVals) - ls
- max1, thr1 = MIN_JUMP, 255
- for i in range(ls, l):
- jump = QVals[i + ls] - QVals[i - ls]
- if(jump > max1):
- max1 = jump
- thr1 = QVals[i - ls] + jump / 2
-
-# NOTE: thr2 is deprecated, thus is JUMP_DELTA
- # Make use of the fact that the JUMP_DELTA(Vertical gap ofc) between
- # values at detected jumps would be atleast 20
- max2, thr2 = MIN_JUMP, 255
- # Requires atleast 1 gray box to be present (Roll field will ensure this)
- for i in range(ls, l):
- jump = QVals[i + ls] - QVals[i - ls]
- newThr = QVals[i - ls] + jump / 2
- if(jump > max2 and abs(thr1 - newThr) > JUMP_DELTA):
- max2 = jump
- thr2 = newThr
- # globalTHR = min(thr1,thr2)
- globalTHR, j_low, j_high = thr1, thr1 - max1 // 2, thr1 + max1 // 2
-
- # # For normal images
- # thresholdRead = 116
- # if(thr1 > thr2 and thr2 > thresholdRead):
- # print("Note: taking safer thr line.")
- # globalTHR, j_low, j_high = thr2, thr2 - max2//2, thr2 + max2//2
-
- if(plotTitle is not None):
- f, ax = plt.subplots()
- ax.bar(range(len(QVals_orig)), QVals if sortInPlot else QVals_orig)
- ax.set_title(plotTitle)
- thrline = ax.axhline(globalTHR, color='green', ls='--', linewidth=5)
- thrline.set_label("Global Threshold")
- thrline = ax.axhline(thr2, color='red', ls=':', linewidth=3)
- thrline.set_label("THR2 Line")
- # thrline=ax.axhline(j_low,color='red',ls='-.', linewidth=3)
- # thrline=ax.axhline(j_high,color='red',ls='-.', linewidth=3)
- # thrline.set_label("Boundary Line")
- # ax.set_ylabel("Mean Intensity")
- ax.set_ylabel("Values")
- ax.set_xlabel("Position")
- ax.legend()
- if(plotShow):
- plt.title(plotTitle)
- plt.show()
-
- return globalTHR, j_low, j_high
-
-
-def getLocalThreshold(
- qNo,
- QVals,
- globalTHR,
- noOutliers,
- plotTitle=None,
- plotShow=True):
- """
- TODO: Update this documentation too-
- //No more - Assumption : Colwise background color is uniformly gray or white, but not alternating. In this case there is atmost one jump.
-
- 0 Jump :
- <-- safe THR?
- .......
- ...|||||||
- |||||||||| <-- safe THR?
- // How to decide given range is above or below gray?
- -> global QVals shall absolutely help here. Just run same function on total QVals instead of colwise _//
- How to decide it is this case of 0 jumps
-
- 1 Jump :
- ......
- ||||||
- |||||| <-- risky THR
- |||||| <-- safe THR
- ....||||||
- ||||||||||
-
- """
- # Sort the Q vals
- QVals = sorted(QVals)
-
- # Small no of pts cases:
- # base case: 1 or 2 pts
- if(len(QVals) < 3):
- thr1 = globalTHR if np.max(
- QVals) - np.min(QVals) < MIN_GAP else np.mean(QVals)
- else:
- # qmin, qmax, qmean, qstd = round(np.min(QVals),2), round(np.max(QVals),2), round(np.mean(QVals),2), round(np.std(QVals),2)
- # GVals = [round(abs(q-qmean),2) for q in QVals]
- # gmean, gstd = round(np.mean(GVals),2), round(np.std(GVals),2)
- # # DISCRETION: Pretty critical factor in reading response
- # # Doesn't work well for small number of values.
- # DISCRETION = 2.7 # 2.59 was closest hit, 3.0 is too far
- # L2MaxGap = round(max([abs(g-gmean) for g in GVals]),2)
- # if(L2MaxGap > DISCRETION*gstd):
- # noOutliers = False
-
- # # ^Stackoverflow method
- # print(qNo, noOutliers,"qstd",round(np.std(QVals),2), "gstd", gstd,"Gaps in gvals",sorted([round(abs(g-gmean),2) for g in GVals],reverse=True), '\t',round(DISCRETION*gstd,2), L2MaxGap)
-
- # else:
- # Find the LARGEST GAP and set it as threshold: //(FIRST LARGE GAP)
- l = len(QVals) - 1
- max1, thr1 = MIN_JUMP, 255
- for i in range(1, l):
- jump = QVals[i + 1] - QVals[i - 1]
- if(jump > max1):
- max1 = jump
- thr1 = QVals[i - 1] + jump / 2
- # print(qNo,QVals,max1)
-
- # If not confident, then only take help of globalTHR
- if(max1 < CONFIDENT_JUMP):
- if(noOutliers):
- # All Black or All White case
- thr1 = globalTHR
- else:
- # TODO: Low confidence parameters here
- pass
-
- # if(thr1 == 255):
- # print("Warning: threshold is unexpectedly 255! (Outlier Delta issue?)",plotTitle)
-
- if(plotShow and plotTitle is not None):
- f, ax = plt.subplots()
- ax.bar(range(len(QVals)), QVals)
- thrline = ax.axhline(thr1, color='green', ls=('-.'), linewidth=3)
- thrline.set_label("Local Threshold")
- thrline = ax.axhline(globalTHR, color='red', ls=':', linewidth=5)
- thrline.set_label("Global Threshold")
- ax.set_title(plotTitle)
- ax.set_ylabel("Bubble Mean Intensity")
- ax.set_xlabel("Bubble Number(sorted)")
- ax.legend()
- # TODO append QStrip to this plot-
- # appendSaveImg(6,getPlotImg())
- if(plotShow):
- plt.show()
- return thr1
-
-# from matplotlib.ticker import MaxNLocator
-# def plotArray(QVals, plotTitle, sort = False, plot=True ):
-# f, ax = plt.subplots()
-# if(sort):
-# QVals = sorted(QVals)
-# ax.bar(range(len(QVals)),QVals)
-# ax.set_title(plotTitle)
-# ax.set_ylabel("Values")
-# ax.set_xlabel("Position")
-# ax.xaxis.set_major_locator(MaxNLocator(integer=True))
-# if(plot):
-# plt.show()
-# # else: they will call this
-# # appendSaveImg(appendImgLvl,getPlotImg())
-
-
-def saveImg(path, final_marked):
- print('Saving Image to ' + path)
- cv2.imwrite(path, final_marked)
-
-
-def readResponse(template, image, name, savedir=None, autoAlign=False):
- global clahe
-
- try:
- img = image.copy()
- origDim = img.shape[:2]
- # print("noCropping dim", origDim)
- img = resize_util(img, template.dims[0], template.dims[1])
- # print("Resized dim", img.shape[:2])
-
- if(img.max() > img.min()):
- img = normalize_util(img)
- # Processing copies
- transp_layer = img.copy()
- final_marked = img.copy()
- # putLabel(final_marked,"Crop Size: " + str(origDim[0])+"x"+str(origDim[1]) + " "+name, size=1)
-
- morph = img.copy()
- appendSaveImg(3, morph)
-
- # TODO: evaluate if CLAHE is really req
- if(autoAlign):
- # Note: clahe is good for morphology, bad for thresholding
- morph = clahe.apply(morph)
- appendSaveImg(3, morph)
- # Remove shadows further, make columns/boxes darker (less gamma)
- morph = adjust_gamma(morph, GAMMA_LOW)
- ret, morph = cv2.threshold(morph, 220, 220, cv2.THRESH_TRUNC)
- morph = normalize_util(morph)
- appendSaveImg(3, morph)
- if(showimglvl >= 4):
- show("morph1", morph, 0, 1)
-
- # Overlay Transparencies
- alpha = 0.65
- alpha1 = 0.55
-
- boxW, boxH = template.bubbleDims
- lang = ['E', 'H']
- OMRresponse = {}
-
- multimarked, multiroll = 0, 0
-
- # TODO Make this part useful for visualizing status checks
- # blackVals=[0]
- # whiteVals=[255]
-
- if(showimglvl >= 5):
- # "QTYPE_ROLL":[]}#,"QTYPE_MED":[]}
- allCBoxvals = {"Int": [], "Mcq": []}
- # ,"QTYPE_ROLL":[]}#,"QTYPE_MED":[]}
- qNums = {"Int": [], "Mcq": []}
-
- # Find Shifts for the QBlocks --> Before calculating threshold!
- if(autoAlign):
- # print("Begin Alignment")
- # Open : erode then dilate
- # Vertical kernel
- v_kernel = cv2.getStructuringElement(cv2.MORPH_RECT, (2, 10))
- morph_v = cv2.morphologyEx(
- morph, cv2.MORPH_OPEN, v_kernel, iterations=3)
- ret, morph_v = cv2.threshold(morph_v, 200, 200, cv2.THRESH_TRUNC)
- morph_v = 255 - normalize_util(morph_v)
-
- if(showimglvl >= 3):
- show("morphed_vertical", morph_v, 0, 1)
-
- # show("morph1",morph,0,1)
- # show("morphed_vertical",morph_v,0,1)
-
- appendSaveImg(3, morph_v)
-
- morphTHR = 60 # for Mobile images
- # morphTHR = 40 # for scan Images
- # best tuned to 5x5 now
- _, morph_v = cv2.threshold(
- morph_v, morphTHR, 255, cv2.THRESH_BINARY)
- morph_v = cv2.erode(
- morph_v, np.ones(
- (5, 5), np.uint8), iterations=2)
-
- appendSaveImg(3, morph_v)
- # h_kernel = cv2.getStructuringElement(cv2.MORPH_RECT, (10, 2))
- # morph_h = cv2.morphologyEx(morph, cv2.MORPH_OPEN, h_kernel, iterations=3)
- # ret, morph_h = cv2.threshold(morph_h,200,200,cv2.THRESH_TRUNC)
- # morph_h = 255 - normalize_util(morph_h)
- # show("morph_h",morph_h,0,1)
- # _, morph_h = cv2.threshold(morph_h,morphTHR,255,cv2.THRESH_BINARY)
- # morph_h = cv2.erode(morph_h, np.ones((5,5),np.uint8), iterations = 2)
- if(showimglvl >= 3):
- show("morph_thr_eroded", morph_v, 0, 1)
-
- appendSaveImg(6, morph_v)
-
- # template alignment code
- # OUTPUT : each QBlock.shift is updated
- for QBlock in template.QBlocks:
- s, d = QBlock.orig, QBlock.dims
- # internal constants - wont need change much
- # TODO - ALIGN_STRIDE would depend on template's Dimensions
- ALIGN_STRIDE, MATCH_COL, ALIGN_STEPS = 1, 5, int(boxW * 2 / 3)
- shift, steps = 0, 0
- THK = 3
- while steps < ALIGN_STEPS:
- L = np.mean(morph_v[s[1]:s[1]+d[1],s[0]+shift-THK:-THK+s[0]+shift+MATCH_COL])
- R = np.mean(morph_v[s[1]:s[1]+d[1],s[0]+shift-MATCH_COL+d[0]+THK:THK+s[0]+shift+d[0]])
-
- # For demonstration purposes-
- if(QBlock.key == "Int1"):
- ret = morph_v.copy()
- cv2.rectangle(ret,(s[0]+shift-THK,s[1]),(s[0]+shift+THK+d[0],s[1]+d[1]),CLR_WHITE,3)
- appendSaveImg(6, ret)
- # print(shift, L, R)
- LW, RW = L > 100, R > 100
- if(LW):
- if(RW):
- break
- else:
- shift -= ALIGN_STRIDE
- else:
- if(RW):
- shift += ALIGN_STRIDE
- else:
- break
- steps += 1
-
- QBlock.shift = shift
- # print("Aligned QBlock: ",QBlock.key,"Corrected Shift:", QBlock.shift,", Dimensions:", QBlock.dims, "orig:", QBlock.orig,'\n')
- # print("End Alignment")
-
- final_align = None
- if(showimglvl >= 2):
- initial_align = drawTemplateLayout(img, template, shifted=False)
- final_align = drawTemplateLayout(
- img, template, shifted=True, draw_qvals=True)
- # appendSaveImg(4,mean_vals)
- appendSaveImg(2, initial_align)
- appendSaveImg(2, final_align)
- appendSaveImg(5, img)
- if(autoAlign):
- final_align = np.hstack((initial_align, final_align))
-
- # Get mean vals n other stats
- allQVals, allQStripArrs, allQStdVals = [], [], []
- totalQStripNo = 0
- for QBlock in template.QBlocks:
- QStdVals = []
- for qStrip, qBoxPts in QBlock.traverse_pts:
- QStripvals = []
- for pt in qBoxPts:
- # shifted
- x, y = (pt.x + QBlock.shift, pt.y)
- rect = [y, y + boxH, x, x + boxW]
- QStripvals.append(
- cv2.mean(img[rect[0]:rect[1], rect[2]:rect[3]])[0])
- QStdVals.append(round(np.std(QStripvals), 2))
- allQStripArrs.append(QStripvals)
- # _, _, _ = getGlobalThreshold(QStripvals, "QStrip Plot", plotShow=False, sortInPlot=True)
- # hist = getPlotImg()
- # show("QStrip "+qBoxPts[0].qNo, hist, 0, 1)
- allQVals.extend(QStripvals)
- # print(totalQStripNo, qBoxPts[0].qNo, QStdVals[len(QStdVals)-1])
- totalQStripNo += 1
- allQStdVals.extend(QStdVals)
- # print("Begin getGlobalThresholdStd")
- globalStdTHR, jstd_low, jstd_high = getGlobalThreshold(allQStdVals)# , "Q-wise Std-dev Plot", plotShow=True, sortInPlot=True)
- # print("End getGlobalThresholdStd")
- # print("Begin getGlobalThreshold")
- # plt.show()
- # hist = getPlotImg()
- # show("StdHist", hist, 0, 1)
-
- # Note: Plotting takes Significant times here --> Change Plotting args
- # to support showimglvl
- # , "Mean Intensity Histogram",plotShow=True, sortInPlot=True)
- globalTHR, j_low, j_high = getGlobalThreshold(allQVals, looseness=4)
-
- # TODO colorama
- print(
- "Thresholding:\t\t globalTHR: ", round(
- globalTHR, 2), "\tglobalStdTHR: ", round(
- globalStdTHR, 2), "\t(Looks like a Xeroxed OMR)" if(
- globalTHR == 255) else "")
- # plt.show()
- # hist = getPlotImg()
- # show("StdHist", hist, 0, 1)
-
- # print("End getGlobalThreshold")
-
- # if(showimglvl>=1):
- # hist = getPlotImg()
- # show("Hist", hist, 0, 1)
- # appendSaveImg(4,hist)
- # appendSaveImg(5,hist)
- # appendSaveImg(2,hist)
-
- perOMRThresholdAvg, totalQStripNo, totalQBoxNo = 0, 0, 0
- for QBlock in template.QBlocks:
- blockQStripNo = 1 # start from 1 is fine here
- shift = QBlock.shift
- s, d = QBlock.orig, QBlock.dims
- key = QBlock.key[:3]
- # cv2.rectangle(final_marked,(s[0]+shift,s[1]),(s[0]+shift+d[0],s[1]+d[1]),CLR_BLACK,3)
- for qStrip, qBoxPts in QBlock.traverse_pts:
- # All Black or All White case
- noOutliers = allQStdVals[totalQStripNo] < globalStdTHR
- # print(totalQStripNo, qBoxPts[0].qNo, allQStdVals[totalQStripNo], "noOutliers:", noOutliers)
- perQStripThreshold = getLocalThreshold(qBoxPts[0].qNo, allQStripArrs[totalQStripNo],
- globalTHR, noOutliers,
- "Mean Intensity Histogram for " + key + "." +
- qBoxPts[0].qNo + '.' +
- str(blockQStripNo),
- showimglvl >= 6)
- # print(qBoxPts[0].qNo,key,blockQStripNo, "THR: ",round(perQStripThreshold,2))
- perOMRThresholdAvg += perQStripThreshold
-
- # Note: Little debugging visualization - view the particular Qstrip
- # if(
- # 0
- # # or "q17" in (qBoxPts[0].qNo)
- # # or (qBoxPts[0].qNo+str(blockQStripNo))=="q15"
- # ):
- # st, end = qStrip
- # show("QStrip: "+key+"-"+str(blockQStripNo), img[st[1] : end[1], st[0]+shift : end[0]+shift],0)
-
- for pt in qBoxPts:
- # shifted
- x, y = (pt.x + QBlock.shift, pt.y)
- boxval0 = allQVals[totalQBoxNo]
- detected = perQStripThreshold > boxval0
-
- # TODO: add an option to select PLUS SIGN RESPONSE READING
- # extra_check_rects = []
- # # [y,y+boxH,x,x+boxW]
- # for rect in extra_check_rects:
- # # Note: This is NOT pixel-based thresholding, It is boxed mean-thresholding
- # boxval = cv2.mean(img[ rect[0]:rect[1] , rect[2]:rect[3] ])[0]
- # if(perQStripThreshold > boxval):
- # # for critical analysis
- # boxval0 = max(boxval,boxval0)
- # detected=True
- # break
-
- if (detected):
- cv2.rectangle(final_marked,(int(x+boxW/12),int(y+boxH/12)),(int(x+boxW-boxW/12),int(y+boxH-boxH/12)), CLR_DARK_GRAY, 3)
- else:
- cv2.rectangle(final_marked,(int(x+boxW/10),int(y+boxH/10)),(int(x+boxW-boxW/10),int(y+boxH-boxH/10)), CLR_GRAY,-1)
-
- # TODO Make this part useful! (Abstract visualizer to check
- # status)
- if (detected):
- q, val = pt.qNo, str(pt.val)
- cv2.putText(
- final_marked, val, (x, y), cv2.FONT_HERSHEY_SIMPLEX, TEXT_SIZE, (20, 20, 10), int(
- 1 + 3.5 * TEXT_SIZE))
- # Only send rolls multi-marked in the directory
- multimarkedL = q in OMRresponse
- multimarked = multimarkedL or multimarked
- OMRresponse[q] = (
- OMRresponse[q] + val) if multimarkedL else val
- multiroll = multimarkedL and 'roll' in str(q)
- # blackVals.append(boxval0)
- # else:
- # whiteVals.append(boxval0)
-
- totalQBoxNo += 1
- # /for qBoxPts
- # /for qStrip
-
- if(showimglvl >= 5):
- if(key in allCBoxvals):
- qNums[key].append(key[:2] + '_c' + str(blockQStripNo))
- allCBoxvals[key].append(allQStripArrs[totalQStripNo])
-
- blockQStripNo += 1
- totalQStripNo += 1
- # /for QBlock
-
- # TODO: move this validation into template.py -
- if(totalQStripNo == 0):
- print(
- "\n\t UNEXPECTED Template Incorrect Error: totalQStripNo is zero! QBlocks: ",
- template.QBlocks)
- exit(21)
-
- perOMRThresholdAvg /= totalQStripNo
- perOMRThresholdAvg = round(perOMRThresholdAvg, 2)
- # Translucent
- cv2.addWeighted(
- final_marked,
- alpha,
- transp_layer,
- 1 - alpha,
- 0,
- final_marked)
- # Box types
- if(showimglvl >= 5):
- # plt.draw()
- f, axes = plt.subplots(len(allCBoxvals), sharey=True)
- f.canvas.set_window_title(name)
- ctr = 0
- typeName = {
- "Int": "Integer",
- "Mcq": "MCQ",
- "Med": "MED",
- "Rol": "Roll"}
- for k, boxvals in allCBoxvals.items():
- axes[ctr].title.set_text(typeName[k] + " Type")
- axes[ctr].boxplot(boxvals)
- # thrline=axes[ctr].axhline(perOMRThresholdAvg,color='red',ls='--')
- # thrline.set_label("Average THR")
- axes[ctr].set_ylabel("Intensity")
- axes[ctr].set_xticklabels(qNums[k])
- # axes[ctr].legend()
- ctr += 1
- # imshow will do the waiting
- plt.tight_layout(pad=0.5)
- plt.show()
-
- if(showimglvl >= 3 and final_align is not None):
- final_align = resize_util_h(final_align, int(display_height))
- # [final_align.shape[1],0])
- show("Template Alignment Adjustment", final_align, 0, 0)
-
- # TODO: refactor "type(savedir) != type(None) "
- if (saveMarked and type(savedir) != type(None)):
- if(multiroll):
- savedir = savedir + '_MULTI_/'
- saveImg(savedir + name, final_marked)
-
- if(showimglvl >= 1):
- show("Final Marked Bubbles : " + name,
- resize_util_h(final_marked, int(display_height * 1.3)), 1, 1)
-
- appendSaveImg(2, final_marked)
-
- for i in range(saveimglvl):
- saveOrShowStacks(i + 1, name, savedir)
-
- return OMRresponse, final_marked, multimarked, multiroll
-
- except Exception as e:
- exc_type, exc_obj, exc_tb = sys.exc_info()
- fname = os.path.split(exc_tb.tb_frame.f_code.co_filename)[1]
- print("Error from readResponse: ", e)
- print(exc_type, fname, exc_tb.tb_lineno)
-
-
-def saveOrShowStacks(key, name, savedir=None, pause=1):
- global saveImgList
- if(saveimglvl >= int(key) and saveImgList[key] != []):
- result = np.hstack(
- tuple([resize_util_h(img, uniform_height) for img in saveImgList[key]]))
- result = resize_util(result,
- min(len(saveImgList[key]) * uniform_width // 3,
- int(uniform_width * 2.5)))
- if (type(savedir) != type(None)):
- saveImg(savedir+'stack/'+name+'_'+str(key)+'_stack.jpg', result)
- else:
- show(name + '_' + str(key), result, pause, 0)
[](https://www.paypal.me/Udayraj123/500)
+
+_Find OMRChecker on_ [**_Product Hunt_**](https://www.producthunt.com/posts/omr-checker/) **|** [**_Reddit_**](https://www.reddit.com/r/computervision/comments/ccbj6f/omrchecker_grade_exams_using_python_and_opencv/) **|** [**Discord**](https://discord.gg/qFv2Vqf) **|** [**Linkedin**](https://www.linkedin.com/pulse/open-source-talks-udayraj-udayraj-deshmukh/) **|** [**goodfirstissue.dev**](https://goodfirstissue.dev/language/python) **|** [**codepeak.tech**](https://www.codepeak.tech/) **|** [**fossoverflow.dev**](https://fossoverflow.dev/projects) **|** [**Interview on Console by CodeSee**](https://console.substack.com/p/console-140) **|** [**Open Source Hub**](https://opensourcehub.io/udayraj123/omrchecker)
-
-*Find OMRChecker on* [***Product Hunt***](https://www.producthunt.com/posts/omr-checker/) **|** [***Hacker News***](https://news.ycombinator.com/item?id=20420602) **|** [***Reddit***](https://www.reddit.com/r/computervision/comments/ccbj6f/omrchecker_grade_exams_using_python_and_opencv/) **|** [***Swyya***](https://www.swyya.com/projects/omrchecker) **|** [](https://discord.gg/qFv2Vqf)
+
+
diff --git a/docs/assets/colored_output.jpg b/docs/assets/colored_output.jpg
new file mode 100644
index 00000000..3cafa473
Binary files /dev/null and b/docs/assets/colored_output.jpg differ
diff --git a/globals.py b/globals.py
deleted file mode 100644
index 5f9cd929..00000000
--- a/globals.py
+++ /dev/null
@@ -1,140 +0,0 @@
-"""
-
-Designed and Developed by-
-Udayraj Deshmukh
-https://github.com/Udayraj123
-
-"""
-
-"""
-Constants
-"""
-display_height = int(480)
-display_width = int(640)
-windowWidth = 1280
-windowHeight = 720
-
-saveMarked = 1
-saveCropped = 1
-showimglvl = 4
-saveimglvl = 0
-PRELIM_CHECKS = 0
-saveImgList = {}
-resetpos = [0, 0]
-explain = 0
-# autorotate=1
-
-BATCH_NO = 1000
-NO_MARKER_ERR = 12
-MULTI_BUBBLE_WARN = 15
-
-# name of template file
-TEMPLATE_FILE = 'template.json'
-MARKER_FILE = "omr_marker.jpg"
-
-# For preProcessing
-GAMMA_LOW = 0.7
-GAMMA_HIGH = 1.25
-
-ERODE_SUB_OFF = 1
-
-# For new ways of determining threshold
-MIN_GAP, MIN_STD = 30, 25
-MIN_JUMP = 25
-# If only not confident, take help of globalTHR
-CONFIDENT_JUMP = MIN_JUMP + 15
-JUMP_DELTA = 30
-# MIN_GAP : worst case gap of black and gray
-
-# Templ alignment parameters
-ALIGN_RANGE = range(-5, 6, 1)
-# TODO ^THIS SHOULD BE IN LAYOUT FILE AS ITS RELATED TO DIMENSIONS
-# ALIGN_RANGE = [-6,-4,-2,-1,0,1,2,4,6]
-
-# max threshold difference for template matching
-thresholdVar = 0.41
-
-# TODO: remove unnec variables here-
-thresholdCircle = 0.3
-marker_rescale_range = (35, 100)
-marker_rescale_steps = 10
-
-# Presentation variables
-uniform_height = int(1231 / 1.5)
-uniform_width = int(1000 / 1.5)
-# Original dims are about (3527, 2494)
-
-# Any input images should be resized to this--
-uniform_width_hd = int(uniform_width * 1.5)
-uniform_height_hd = int(uniform_height * 1.5)
-
-TEXT_SIZE = 0.95
-CLR_BLACK = (50, 150, 150)
-CLR_WHITE = (250, 250, 250)
-CLR_GRAY = (130, 130, 130)
-# CLR_DARK_GRAY = (190,190,190)
-CLR_DARK_GRAY = (100, 100, 100)
-
-MIN_PAGE_AREA = 80000
-
-# Filepaths
-
-
-class Paths:
- def __init__(self, output):
- self.output = output
- self.saveMarkedDir = f'{output}/CheckedOMRs/'
- self.resultDir = f'{output}/Results/'
- self.manualDir = f'{output}/Manual/'
- self.errorsDir = f'{self.manualDir}ErrorFiles/'
- self.badRollsDir = f'{self.manualDir}BadRollNosFiles/'
- self.multiMarkedDir = f'{self.manualDir}MultiMarkedFiles/'
-
-
-"""
-Variables
-"""
-filesMoved = 0
-filesNotMoved = 0
-
-# for positioning image windows
-windowX, windowY = 0, 0
-
-
-# TODO: move to template or similar json
-Answers = {
- 'J': {
- 'q1': ['B'], 'q2': ['B'], 'q3': ['B'], 'q4': ['C'], 'q5': ['0', '00'], 'q6': ['0', '00'], 'q7': ['4', '04'],
- 'q8': ['9', '09'], 'q9': ['11', '11'], 'q10': ['C'], 'q11': ['C'], 'q12': ['B'], 'q13': ['C'],
- 'q14': ['C'], 'q15': ['B'], 'q16': ['C'], 'q17': ['BONUS'], 'q18': ['A'], 'q19': ['C'], 'q20': ['B']},
- 'H': {
- 'q1': ['B'], 'q2': ['BONUS'], 'q3': ['A'], 'q4': ['B'], 'q5': ['A'], 'q6': ['B'], 'q7': ['B'],
- 'q8': ['C'], 'q9': ['4', '04'], 'q10': ['4', '04'], 'q11': ['5', '05'], 'q12': ['1', '01'], 'q13': ['28'],
- 'q14': ['C'], 'q15': ['B'], 'q16': ['C'], 'q17': ['C'], 'q18': ['C'], 'q19': ['B'], 'q20': ['C']},
- 'JK': {
- 'q1': ['B'], 'q2': ['B'], 'q3': ['B'], 'q4': ['C'], 'q5': ['0', '00'], 'q6': ['0', '00'], 'q7': ['4', '04'],
- 'q8': ['9', '09'], 'q9': ['11', '11'], 'q10': ['C'], 'q11': ['C'], 'q12': ['B'], 'q13': ['C'],
- 'q14': ['C'], 'q15': ['B'], 'q16': ['C'], 'q17': ['BONUS'], 'q18': ['A'], 'q19': ['C'], 'q20': ['B']},
- 'HK': {
- 'q1': ['B'], 'q2': ['BONUS'], 'q3': ['A'], 'q4': ['B'], 'q5': ['B'], 'q6': ['B'], 'q7': ['B'],
- 'q8': ['C'], 'q9': ['4', '04'], 'q10': ['4', '04'], 'q11': ['5', '05'], 'q12': ['1', '01'], 'q13': ['28'],
- 'q14': ['C'], 'q15': ['B'], 'q16': ['C'], 'q17': ['C'], 'q18': ['C'], 'q19': ['B'], 'q20': ['C']},
-}
-
-# TODO: Make this generalized and move it to samples
-Sections = {
- 'J': {
- 'Fibo1': {'ques': [1, 2, 3, 4], '+seq': [2, 3, 5, 8], '-seq': [0, 1, 1, 2]},
- 'Power1': {'ques': [5, 6, 7, 8, 9], '+seq': [1, 2, 4, 8, 16], '-seq': [0, 0, 0, 0, 0]},
- 'Fibo2': {'ques': [10, 11, 12, 13], '+seq': [2, 3, 5, 8], '-seq': [0, 1, 1, 2]},
- 'allNone1': {'ques': [14, 15, 16], 'marks': 12},
- 'Boom1': {'ques': [17, 18, 19, 20], '+seq': [3, 3, 3, 3], '-seq': [1, 1, 1, 1]},
- },
- 'H': {
- 'Boom1': {'ques': [1, 2, 3, 4], '+seq': [3, 3, 3, 3], '-seq': [1, 1, 1, 1]},
- 'Fibo1': {'ques': [5, 6, 7, 8], '+seq': [2, 3, 5, 8], '-seq': [0, 1, 1, 2]},
- 'Power1': {'ques': [9, 10, 11, 12, 13], '+seq': [1, 2, 4, 8, 16], '-seq': [0, 0, 0, 0, 0]},
- 'allNone1': {'ques': [14, 15, 16], 'marks': 12},
- 'Boom2': {'ques': [17, 18, 19, 20], '+seq': [3, 3, 3, 3], '-seq': [1, 1, 1, 1]},
- },
-}
diff --git a/main.py b/main.py
index eac70a50..dfecbca3 100644
--- a/main.py
+++ b/main.py
@@ -1,546 +1,99 @@
"""
-Designed and Developed by-
-Udayraj Deshmukh
-https://github.com/Udayraj123
+ OMRChecker
+
+ Author: Udayraj Deshmukh
+ Github: https://github.com/Udayraj123
"""
-import re
-import os
-import cv2
import argparse
-import numpy as np
-import pandas as pd
-import matplotlib.pyplot as plt
-
-# from utils import * #Now imported via template
-from globals import *
-from template import *
-from glob import glob
-from csv import QUOTE_NONNUMERIC
-from time import localtime, strftime, time
-
-
-# TODO(beginner task) :-
-# from colorama import init
-# init()
-# from colorama import Fore, Back, Style
-
-def process_dir(root_dir, subdir, template):
- curr_dir = os.path.join(root_dir, subdir)
-
- # Look for template in current dir
- template_file = os.path.join(curr_dir, TEMPLATE_FILE)
- if os.path.exists(template_file):
- template = Template(template_file)
-
- # look for images in current dir to process
- paths = Paths(os.path.join(args['output_dir'], subdir))
- exts = ('*.png', '*.jpg')
- omr_files = sorted(
- [f for ext in exts for f in glob(os.path.join(curr_dir, ext))])
-
- # Exclude marker image if exists
- if(template and template.marker_path):
- omr_files = [f for f in omr_files if f != template.marker_path]
-
- subfolders = sorted([file for file in os.listdir(
- curr_dir) if os.path.isdir(os.path.join(curr_dir, file))])
- if omr_files:
- args_local = args.copy()
- if("OverrideFlags" in template.options):
- args_local.update(template.options["OverrideFlags"])
- print('\n------------------------------------------------------------------')
- print(f'Processing directory "{curr_dir}" with settings- ')
- print("\tTotal images : %d" % (len(omr_files)))
- print("\tCropping Enabled : " + str(not args_local["noCropping"]))
- print("\tAuto Alignment : " + str(args_local["autoAlign"]))
- print("\tUsing Template : " + str(template.path) if(template) else "N/A")
- print("\tUsing Marker : " + str(template.marker_path)
- if(template.marker is not None) else "N/A")
- print('')
-
- if not template:
- print(f'Error: No template file when processing {curr_dir}.')
- print(
- f' Place {TEMPLATE_FILE} in the directory or specify a template using -t.')
- return
-
- setup_dirs(paths)
- output_set = setup_output(paths, template)
- process_files(omr_files, template, args_local, output_set)
- elif(len(subfolders) == 0):
- # the directory should have images or be non-leaf
- print(f'Note: No valid images or subfolders found in {curr_dir}')
-
- # recursively process subfolders
- for folder in subfolders:
- process_dir(root_dir, os.path.join(subdir, folder), template)
-
-
-def checkAndMove(error_code, filepath, filepath2):
- # print("Dummy Move: "+filepath, " --> ",filepath2)
- global filesNotMoved
- filesNotMoved += 1
- return True
-
- global filesMoved
- if(not os.path.exists(filepath)):
- print('File already moved')
- return False
- if(os.path.exists(filepath2)):
- print('ERROR : Duplicate file at ' + filepath2)
- return False
-
- print("Moved: " + filepath, " --> ", filepath2)
- os.rename(filepath, filepath2)
- filesMoved += 1
- return True
-
-
-def processOMR(template, omrResp):
- # Note: This is a reference function. It is not part of the OMR checker
- # So its implementation is completely subjective to user's requirements.
- csvResp = {}
-
- # symbol for absent response
- UNMARKED_SYMBOL = ''
-
- # print("omrResp",omrResp)
-
- # Multi-column/multi-row questions which need to be concatenated
- for qNo, respKeys in template.concats.items():
- csvResp[qNo] = ''.join([omrResp.get(k, UNMARKED_SYMBOL)
- for k in respKeys])
-
- # Single-column/single-row questions
- for qNo in template.singles:
- csvResp[qNo] = omrResp.get(qNo, UNMARKED_SYMBOL)
-
- # Note: Concatenations and Singles together should be mutually exclusive
- # and should cover all questions in the template(exhaustive)
- # TODO: ^add a warning if omrResp has unused keys remaining
- return csvResp
-
-
-def report(
- Status,
- streak,
- scheme,
- qNo,
- marked,
- ans,
- prevmarks,
- currmarks,
- marks):
- print(
- '%s \t %s \t\t %s \t %s \t %s \t %s \t %s ' % (qNo,
- Status,
- str(streak),
- '[' + scheme + '] ',
- (str(prevmarks) + ' + ' + str(currmarks) + ' =' + str(marks)),
- str(marked),
- str(ans)))
-
-# check sectionwise only.
-
-
-def evaluate(resp, squad="H", explain=False):
- # TODO: @contributors - Need help generalizing this function
- global Answers, Sections
- marks = 0
- answers = Answers[squad]
- if(explain):
- print('Question\tStatus \t Streak\tSection \tMarks_Update\tMarked:\tAnswer:')
- for scheme, section in Sections[squad].items():
- sectionques = section['ques']
- prevcorrect = None
- allflag = 1
- streak = 0
- for q in sectionques:
- qNo = 'q' + str(q)
- ans = answers[qNo]
- marked = resp.get(qNo, 'X')
- firstQ = sectionques[0]
- lastQ = sectionques[len(sectionques) - 1]
- unmarked = marked == 'X' or marked == ''
- bonus = 'BONUS' in ans
- correct = bonus or (marked in ans)
- inrange = 0
-
- if(unmarked or int(q) == firstQ):
- streak = 0
- elif(prevcorrect == correct):
- streak += 1
- else:
- streak = 0
-
- if('allNone' in scheme):
- # loop on all sectionques
- allflag = allflag and correct
- if(q == lastQ):
- # at the end check allflag
- prevcorrect = correct
- currmarks = section['marks'] if allflag else 0
- else:
- currmarks = 0
-
- elif('Proxy' in scheme):
- a = int(ans[0])
- # proximity check
- inrange = 1 if unmarked else (
- float(abs(int(marked) - a)) / float(a) <= 0.25)
- currmarks = section['+marks'] if correct else (
- 0 if inrange else -section['-marks'])
-
- elif('Fibo' in scheme or 'Power' in scheme or 'Boom' in scheme):
- currmarks = section['+seq'][streak] if correct else (
- 0 if unmarked else -section['-seq'][streak])
- elif('TechnoFin' in scheme):
- currmarks = 0
- else:
- print('Invalid Sections')
- prevmarks = marks
- marks += currmarks
-
- if(explain):
- if bonus:
- report('BonusQ', streak, scheme, qNo, marked,
- ans, prevmarks, currmarks, marks)
- elif correct:
- report('Correct', streak, scheme, qNo, marked,
- ans, prevmarks, currmarks, marks)
- elif unmarked:
- report('Unmarked', streak, scheme, qNo, marked,
- ans, prevmarks, currmarks, marks)
- elif inrange:
- report('InProximity', streak, scheme, qNo,
- marked, ans, prevmarks, currmarks, marks)
- else:
- report('Incorrect', streak, scheme, qNo,
- marked, ans, prevmarks, currmarks, marks)
-
- prevcorrect = correct
-
- return marks
-
-
-def setup_output(paths, template):
- ns = argparse.Namespace()
- print("\nChecking Files...")
-
- # Include current output paths
- ns.paths = paths
-
- # custom sort: To use integer order in question names instead of
- # alphabetical - avoids q1, q10, q2 and orders them q1, q2, ..., q10
- ns.respCols = sorted(list(template.concats.keys()) + template.singles,
- key=lambda x: int(x[1:]) if ord(x[1]) in range(48, 58) else 0)
- ns.emptyResp = [''] * len(ns.respCols)
- ns.sheetCols = ['file_id', 'input_path',
- 'output_path', 'score'] + ns.respCols
- ns.OUTPUT_SET = []
- ns.filesObj = {}
- ns.filesMap = {
- "Results": paths.resultDir + 'Results_' + timeNowHrs + '.csv',
- "MultiMarked": paths.manualDir + 'MultiMarkedFiles_.csv',
- "Errors": paths.manualDir + 'ErrorFiles_.csv',
- "BadRollNos": paths.manualDir + 'BadRollNoFiles_.csv'
- }
-
- for fileKey, fileName in ns.filesMap.items():
- if(not os.path.exists(fileName)):
- print("Note: Created new file: %s" % (fileName))
- # still append mode req [THINK!]
- ns.filesObj[fileKey] = open(fileName, 'a')
- # Create Header Columns
- pd.DataFrame([ns.sheetCols], dtype=str).to_csv(
- ns.filesObj[fileKey], quoting=QUOTE_NONNUMERIC, header=False, index=False)
- else:
- print('Present : appending to %s' % (fileName))
- ns.filesObj[fileKey] = open(fileName, 'a')
-
- return ns
-
-
-''' TODO: Refactor into new process flow.
- Currently I have no idea what this does so I left it out'''
-
-
-def preliminary_check():
- filesCounter = 0
- # PRELIM_CHECKS for thresholding
- if(PRELIM_CHECKS):
- # TODO: add more using unit testing
- ALL_WHITE = 255 * \
- np.ones((TEMPLATE.dims[1], TEMPLATE.dims[0]), dtype='uint8')
- OMRresponseDict, final_marked, MultiMarked, multiroll = readResponse(
- ALL_WHITE, name="ALL_WHITE", savedir=None, autoAlign=True)
- print("ALL_WHITE", OMRresponseDict)
- if(OMRresponseDict != {}):
- print("Preliminary Checks Failed.")
- exit(12)
- ALL_BLACK = np.zeros(
- (TEMPLATE.dims[1], TEMPLATE.dims[0]), dtype='uint8')
- OMRresponseDict, final_marked, MultiMarked, multiroll = readResponse(
- ALL_BLACK, name="ALL_BLACK", savedir=None, autoAlign=True)
- print("ALL_BLACK", OMRresponseDict)
- show("Confirm : All bubbles are black", final_marked, 1, 1)
-
-
-def process_files(omr_files, template, args, out):
- start_time = int(time())
- filesCounter = 0
- filesNotMoved = 0
-
- for filepath in omr_files:
- filesCounter += 1
- # For windows filesystem support: all '\' will be replaced by '/'
- filepath = filepath.replace(os.sep, '/')
-
- # Prefixing a 'r' to use raw string (escape character '\' is taken
- # literally)
- finder = re.search(r'.*/(.*)/(.*)', filepath, re.IGNORECASE)
- if(finder):
- inputFolderName, filename = finder.groups()
- else:
- print("Error: Filepath not matching to Regex: " + filepath)
- continue
- # set global var for reading
-
- inOMR = cv2.imread(filepath, cv2.IMREAD_GRAYSCALE)
- print(
- '\n[%d] Processing image: \t' %
- (filesCounter),
- filepath,
- "\tResolution: ",
- inOMR.shape)
-
- OMRCrop = getROI(inOMR, filename, noCropping=args["noCropping"])
-
- if(OMRCrop is None):
- # Error OMR - could not crop
- newfilepath = out.paths.errorsDir + filename
- out.OUTPUT_SET.append([filename] + out.emptyResp)
- if(checkAndMove(NO_MARKER_ERR, filepath, newfilepath)):
- err_line = [filename, filepath,
- newfilepath, "NA"] + out.emptyResp
- pd.DataFrame(
- err_line,
- dtype=str).T.to_csv(
- out.filesObj["Errors"],
- quoting=QUOTE_NONNUMERIC,
- header=False,
- index=False)
- continue
-
- if template.marker is not None:
- OMRCrop = handle_markers(OMRCrop, template.marker, filename)
-
- if(args["setLayout"]):
- templateLayout = drawTemplateLayout(
- OMRCrop, template, shifted=False, border=2)
- show("Template Layout", templateLayout, 1, 1)
- continue
-
- # uniquify
- file_id = inputFolderName + '_' + filename
- savedir = out.paths.saveMarkedDir
- OMRresponseDict, final_marked, MultiMarked, multiroll = \
- readResponse(template, OMRCrop, name=file_id,
- savedir=savedir, autoAlign=args["autoAlign"])
-
- # concatenate roll nos, set unmarked responses, etc
- resp = processOMR(template, OMRresponseDict)
- print("\nRead Response: \t", resp)
-
- # This evaluates and returns the score attribute
- score = evaluate(resp, explain=explain)
- respArray = []
- for k in out.respCols:
- respArray.append(resp[k])
-
- out.OUTPUT_SET.append([filename] + respArray)
-
- # TODO: Add roll number validation here
- if(MultiMarked == 0):
- filesNotMoved += 1
- newfilepath = savedir + file_id
- # Enter into Results sheet-
- results_line = [filename, filepath, newfilepath, score] + respArray
- # Write/Append to results_line file(opened in append mode)
- pd.DataFrame(
- results_line,
- dtype=str).T.to_csv(
- out.filesObj["Results"],
- quoting=QUOTE_NONNUMERIC,
- header=False,
- index=False)
- print("[%d] Graded with score: %.2f" %
- (filesCounter, score), '\t file_id: ', file_id)
- # print(filesCounter,file_id,resp['Roll'],'score : ',score)
- else:
- # MultiMarked file
- print('[%d] MultiMarked, moving File: %s' %
- (filesCounter, file_id))
- newfilepath = out.paths.multiMarkedDir + filename
- if(checkAndMove(MULTI_BUBBLE_WARN, filepath, newfilepath)):
- mm_line = [filename, filepath, newfilepath, "NA"] + respArray
- pd.DataFrame(
- mm_line,
- dtype=str).T.to_csv(
- out.filesObj["MultiMarked"],
- quoting=QUOTE_NONNUMERIC,
- header=False,
- index=False)
- # else:
- # TODO: Add appropriate record handling here
- # pass
-
- # flush after every 20 files for a live view
- if(filesCounter % 20 == 0 or filesCounter == len(omr_files)):
- for fileKey in out.filesMap.keys():
- out.filesObj[fileKey].flush()
-
- timeChecking = round(time() - start_time, 2) if filesCounter else 1
- print('')
- print('Total files moved : %d ' % (filesMoved))
- print('Total files not moved : %d ' % (filesNotMoved))
- print('------------------------------')
- print(
- 'Total files processed : %d (%s)' %
- (filesCounter,
- 'Sum Tallied!' if filesCounter == (
- filesMoved +
- filesNotMoved) else 'Not Tallying!'))
-
- if(showimglvl <= 0):
- print(
- '\nFinished Checking %d files in %.1f seconds i.e. ~%.1f minutes.' %
- (filesCounter, timeChecking, timeChecking / 60))
- print('OMR Processing Rate :\t ~ %.2f seconds/OMR' %
- (timeChecking / filesCounter))
- print('OMR Processing Speed :\t ~ %.2f OMRs/minute' %
- ((filesCounter * 60) / timeChecking))
- else:
- print("\nTotal script time :", timeChecking, "seconds")
-
- if(showimglvl <= 1):
- # TODO: colorama this
- print(
- "\nTip: To see some awesome visuals, open globals.py and increase 'showimglvl'")
-
- evaluate_correctness(template, out)
-
- # Use this data to train as +ve feedback
- if(showimglvl >= 0 and filesCounter > 10):
- # TODO: Find good parameters to plot and depict image set quality
- for x in [thresholdCircles]: # ,badThresholds,veryBadPoints, , mbs]:
- if(x != []):
- x = pd.DataFrame(x)
- print(x.describe())
- plt.plot(range(len(x)), x)
- plt.title("Mystery Plot")
- plt.show()
- else:
- print(x)
-
-
-# Evaluate accuracy based on OMRDataset file generated through moderation
-# portal on the same set of images
-def evaluate_correctness(template, out):
- # TODO: TEST_FILE WOULD BE RELATIVE TO INPUT SUBDIRECTORY NOW-
- TEST_FILE = 'inputs/OMRDataset.csv'
- if(os.path.exists(TEST_FILE)):
- print("\nStarting evaluation for: " + TEST_FILE)
-
- TEST_COLS = ['file_id'] + out.respCols
- y_df = pd.read_csv(
- TEST_FILE, dtype=str)[TEST_COLS].replace(
- np.nan, '', regex=True).set_index('file_id')
-
- if(np.any(y_df.index.duplicated)):
- y_df_filtered = y_df.loc[~y_df.index.duplicated(keep='first')]
- print(
- "WARNING: Found duplicate File-ids in file %s. Removed %d rows from testing data. Rows remaining: %d" %
- (TEST_FILE, y_df.shape[0] - y_df_filtered.shape[0], y_df_filtered.shape[0]))
- y_df = y_df_filtered
-
- x_df = pd.DataFrame(
- out.OUTPUT_SET,
- dtype=str,
- columns=TEST_COLS).set_index('file_id')
- # print("x_df",x_df.head())
- # print("\ny_df",y_df.head())
- intersection = y_df.index.intersection(x_df.index)
-
- # Checking if the merge is okay
- if(intersection.size == x_df.index.size):
- y_df = y_df.loc[intersection]
- x_df['TestResult'] = (x_df == y_df).all(axis=1).astype(int)
- print(x_df.head())
- print("\n\t Accuracy on the %s Dataset: %.6f" %
- (TEST_FILE, (x_df['TestResult'].sum() / x_df.shape[0])))
- else:
- print(
- "\nERROR: Insufficient Testing Data: Have you appended MultiMarked data yet?")
- print("Missing File-ids: ",
- list(x_df.index.difference(intersection)))
-
-
-timeNowHrs = strftime("%I%p", localtime())
-
-# construct the argument parse and parse the arguments
-argparser = argparse.ArgumentParser()
-# https://docs.python.org/3/howto/argparse.html
-# store_true: if the option is specified, assign the value True to
-# args.verbose. Not specifying it implies False.
-argparser.add_argument(
- "-c",
- "--noCropping",
- required=False,
- dest='noCropping',
- action='store_true',
- help="Disables page contour detection - used when page boundary is not visible e.g. document scanner.")
-argparser.add_argument(
- "-a",
- "--autoAlign",
- required=False,
- dest='autoAlign',
- action='store_true',
- help="(experimental) Enables automatic template alignment - use if the scans show slight misalignments.")
-argparser.add_argument(
- "-l",
- "--setLayout",
- required=False,
- dest='setLayout',
- action='store_true',
- help="Set up OMR template layout - modify your json file and run again until the template is set.")
-argparser.add_argument("-i", "--inputDir", required=False, action='append',
- dest='input_dir', help="Specify an input directory.")
-argparser.add_argument("-o", "--outputDir", default='outputs', required=False,
- dest='output_dir', help="Specify an output directory.")
-argparser.add_argument(
- "-t",
- "--template",
- required=False,
- dest='template',
- help="Specify a default template if no template file in input directories.")
-
-
-args, unknown = argparser.parse_known_args()
-args = vars(args)
-if(len(unknown) > 0):
- print("\nError: Unknown arguments:", unknown)
- argparser.print_help()
- exit(11)
-
-if args['template']:
- args['template'] = Template(args['template'])
-
-if args['input_dir'] is None:
- args['input_dir'] = ['inputs']
-
-for root in args['input_dir']:
- process_dir(root, '', args['template'])
+import sys
+from pathlib import Path
+
+from src.entry import entry_point
+from src.logger import logger
+
+
+def parse_args():
+ # construct the argument parse and parse the arguments
+ argparser = argparse.ArgumentParser()
+
+ argparser.add_argument(
+ "-i",
+ "--inputDir",
+ default=["inputs"],
+ # https://docs.python.org/3/library/argparse.html#nargs
+ nargs="*",
+ required=False,
+ type=str,
+ dest="input_paths",
+ help="Specify an input directory.",
+ )
+
+ argparser.add_argument(
+ "-d",
+ "--debug",
+ required=False,
+ dest="debug",
+ action="store_false",
+ help="Enables debugging mode for showing detailed errors",
+ )
+
+ argparser.add_argument(
+ "-o",
+ "--outputDir",
+ default="outputs",
+ required=False,
+ dest="output_dir",
+ help="Specify an output directory.",
+ )
+
+ argparser.add_argument(
+ "-a",
+ "--autoAlign",
+ required=False,
+ dest="autoAlign",
+ action="store_true",
+ help="(experimental) Enables automatic template alignment - \
+ use if the scans show slight misalignments.",
+ )
+
+ argparser.add_argument(
+ "-l",
+ "--setLayout",
+ required=False,
+ dest="setLayout",
+ action="store_true",
+ help="Set up OMR template layout - modify your json file and \
+ run again until the template is set.",
+ )
+
+ (
+ args,
+ unknown,
+ ) = argparser.parse_known_args()
+
+ args = vars(args)
+
+ if len(unknown) > 0:
+ logger.warning(f"\nError: Unknown arguments: {unknown}", unknown)
+ argparser.print_help()
+ exit(11)
+ return args
+
+
+def entry_point_for_args(args):
+ if args["debug"] is True:
+ # Disable tracebacks
+ sys.tracebacklimit = 0
+ for root in args["input_paths"]:
+ entry_point(
+ Path(root),
+ args,
+ )
+
+
+if __name__ == "__main__":
+ args = parse_args()
+ entry_point_for_args(args)
diff --git a/pyproject.toml b/pyproject.toml
new file mode 100644
index 00000000..060309be
--- /dev/null
+++ b/pyproject.toml
@@ -0,0 +1,18 @@
+[tool.black]
+exclude = '''
+(
+ /(
+ \.eggs # exclude a few common directories in the
+ | \.git # root of the project
+ | \.venv
+ | _build
+ | build
+ | dist
+ )/
+ | foo.py # also separately exclude a file named foo.py in
+ # the root of the project
+)
+'''
+include = '\.pyi?$'
+line-length = 88
+target-version = ['py37']
diff --git a/pytest.ini b/pytest.ini
new file mode 100644
index 00000000..84008a21
--- /dev/null
+++ b/pytest.ini
@@ -0,0 +1,6 @@
+# pytest.ini
+[pytest]
+minversion = 7.0
+addopts = -qq --capture=no
+testpaths =
+ src/tests
diff --git a/requirements.dev.txt b/requirements.dev.txt
new file mode 100644
index 00000000..722ab0dd
--- /dev/null
+++ b/requirements.dev.txt
@@ -0,0 +1,7 @@
+-r requirements.txt
+flake8>=6.0.0
+freezegun>=1.2.2
+pre-commit>=3.3.3
+pytest-mock>=3.11.1
+pytest>=7.4.0
+syrupy>=4.0.4
diff --git a/requirements.txt b/requirements.txt
index 505865c9..761d2e51 100644
--- a/requirements.txt
+++ b/requirements.txt
@@ -1,4 +1,8 @@
-imutils>=0.5.2
-matplotlib>=3.0.2
-numpy>=1.16.0
-pandas>=0.24.0
+deepmerge>=1.1.0
+dotmap>=1.3.30
+jsonschema>=4.17.3
+matplotlib>=3.7.1
+numpy>=1.25.0
+pandas>=2.0.2
+rich>=13.4.2
+screeninfo>=0.8.1
diff --git a/samples/answer-key/using-csv/adrian_omr.png b/samples/answer-key/using-csv/adrian_omr.png
new file mode 100644
index 00000000..d8db0994
Binary files /dev/null and b/samples/answer-key/using-csv/adrian_omr.png differ
diff --git a/samples/answer-key/using-csv/answer_key.csv b/samples/answer-key/using-csv/answer_key.csv
new file mode 100644
index 00000000..566201d1
--- /dev/null
+++ b/samples/answer-key/using-csv/answer_key.csv
@@ -0,0 +1,5 @@
+q1,C
+q2,E
+q3,A
+q4,B
+q5,B
\ No newline at end of file
diff --git a/samples/answer-key/using-csv/evaluation.json b/samples/answer-key/using-csv/evaluation.json
new file mode 100644
index 00000000..14a3db25
--- /dev/null
+++ b/samples/answer-key/using-csv/evaluation.json
@@ -0,0 +1,14 @@
+{
+ "source_type": "csv",
+ "options": {
+ "answer_key_csv_path": "answer_key.csv",
+ "should_explain_scoring": true
+ },
+ "marking_schemes": {
+ "DEFAULT": {
+ "correct": "1",
+ "incorrect": "0",
+ "unmarked": "0"
+ }
+ }
+}
diff --git a/samples/answer-key/using-csv/template.json b/samples/answer-key/using-csv/template.json
new file mode 100644
index 00000000..41ec9ffa
--- /dev/null
+++ b/samples/answer-key/using-csv/template.json
@@ -0,0 +1,35 @@
+{
+ "pageDimensions": [
+ 300,
+ 400
+ ],
+ "bubbleDimensions": [
+ 25,
+ 25
+ ],
+ "preProcessors": [
+ {
+ "name": "CropPage",
+ "options": {
+ "morphKernel": [
+ 10,
+ 10
+ ]
+ }
+ }
+ ],
+ "fieldBlocks": {
+ "MCQ_Block_1": {
+ "fieldType": "QTYPE_MCQ5",
+ "origin": [
+ 65,
+ 60
+ ],
+ "fieldLabels": [
+ "q1..5"
+ ],
+ "labelsGap": 52,
+ "bubblesGap": 41
+ }
+ }
+}
diff --git a/samples/answer-key/weighted-answers/evaluation.json b/samples/answer-key/weighted-answers/evaluation.json
new file mode 100644
index 00000000..c0daefcf
--- /dev/null
+++ b/samples/answer-key/weighted-answers/evaluation.json
@@ -0,0 +1,35 @@
+{
+ "source_type": "custom",
+ "options": {
+ "questions_in_order": [
+ "q1..5"
+ ],
+ "answers_in_order": [
+ "C",
+ "E",
+ [
+ "A",
+ "C"
+ ],
+ [
+ [
+ "B",
+ 2
+ ],
+ [
+ "C",
+ "3/2"
+ ]
+ ],
+ "C"
+ ],
+ "should_explain_scoring": true
+ },
+ "marking_schemes": {
+ "DEFAULT": {
+ "correct": "3",
+ "incorrect": "-1",
+ "unmarked": "0"
+ }
+ }
+}
diff --git a/samples/answer-key/weighted-answers/images/adrian_omr.png b/samples/answer-key/weighted-answers/images/adrian_omr.png
new file mode 100644
index 00000000..69a53823
Binary files /dev/null and b/samples/answer-key/weighted-answers/images/adrian_omr.png differ
diff --git a/samples/answer-key/weighted-answers/images/adrian_omr_2.png b/samples/answer-key/weighted-answers/images/adrian_omr_2.png
new file mode 100644
index 00000000..d8db0994
Binary files /dev/null and b/samples/answer-key/weighted-answers/images/adrian_omr_2.png differ
diff --git a/samples/answer-key/weighted-answers/template.json b/samples/answer-key/weighted-answers/template.json
new file mode 100644
index 00000000..41ec9ffa
--- /dev/null
+++ b/samples/answer-key/weighted-answers/template.json
@@ -0,0 +1,35 @@
+{
+ "pageDimensions": [
+ 300,
+ 400
+ ],
+ "bubbleDimensions": [
+ 25,
+ 25
+ ],
+ "preProcessors": [
+ {
+ "name": "CropPage",
+ "options": {
+ "morphKernel": [
+ 10,
+ 10
+ ]
+ }
+ }
+ ],
+ "fieldBlocks": {
+ "MCQ_Block_1": {
+ "fieldType": "QTYPE_MCQ5",
+ "origin": [
+ 65,
+ 60
+ ],
+ "fieldLabels": [
+ "q1..5"
+ ],
+ "labelsGap": 52,
+ "bubblesGap": 41
+ }
+ }
+}
diff --git a/samples/community/Antibodyy/simple_omr_sheet.jpg b/samples/community/Antibodyy/simple_omr_sheet.jpg
new file mode 100644
index 00000000..661d5f4f
Binary files /dev/null and b/samples/community/Antibodyy/simple_omr_sheet.jpg differ
diff --git a/samples/community/Antibodyy/template.json b/samples/community/Antibodyy/template.json
new file mode 100644
index 00000000..7e962bbf
--- /dev/null
+++ b/samples/community/Antibodyy/template.json
@@ -0,0 +1,35 @@
+{
+ "pageDimensions": [
+ 299,
+ 398
+ ],
+ "bubbleDimensions": [
+ 42,
+ 42
+ ],
+ "fieldBlocks": {
+ "MCQBlock1": {
+ "fieldType": "QTYPE_MCQ5",
+ "origin": [
+ 65,
+ 79
+ ],
+ "bubblesGap": 43,
+ "labelsGap": 50,
+ "fieldLabels": [
+ "q1..6"
+ ]
+ }
+ },
+ "preProcessors": [
+ {
+ "name": "CropPage",
+ "options": {
+ "morphKernel": [
+ 10,
+ 10
+ ]
+ }
+ }
+ ]
+}
diff --git a/samples/community/Sandeep-1507/omr-1.png b/samples/community/Sandeep-1507/omr-1.png
new file mode 100644
index 00000000..8c40655e
Binary files /dev/null and b/samples/community/Sandeep-1507/omr-1.png differ
diff --git a/samples/community/Sandeep-1507/omr-2.png b/samples/community/Sandeep-1507/omr-2.png
new file mode 100644
index 00000000..aabef0a5
Binary files /dev/null and b/samples/community/Sandeep-1507/omr-2.png differ
diff --git a/samples/community/Sandeep-1507/omr-3.png b/samples/community/Sandeep-1507/omr-3.png
new file mode 100644
index 00000000..8f1cb5c8
Binary files /dev/null and b/samples/community/Sandeep-1507/omr-3.png differ
diff --git a/samples/community/Sandeep-1507/template.json b/samples/community/Sandeep-1507/template.json
new file mode 100644
index 00000000..b35404e2
--- /dev/null
+++ b/samples/community/Sandeep-1507/template.json
@@ -0,0 +1,234 @@
+{
+ "pageDimensions": [
+ 1189,
+ 1682
+ ],
+ "bubbleDimensions": [
+ 15,
+ 15
+ ],
+ "preProcessors": [
+ {
+ "name": "GaussianBlur",
+ "options": {
+ "kSize": [
+ 3,
+ 3
+ ],
+ "sigmaX": 0
+ }
+ }
+ ],
+ "customLabels": {
+ "Booklet_No": [
+ "b1..7"
+ ]
+ },
+ "fieldBlocks": {
+ "Booklet_No": {
+ "fieldType": "QTYPE_INT",
+ "origin": [
+ 112,
+ 530
+ ],
+ "fieldLabels": [
+ "b1..7"
+ ],
+ "emptyValue": "no",
+ "bubblesGap": 28,
+ "labelsGap": 26.5
+ },
+ "MCQBlock1a1": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q1..10"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 476,
+ 100
+ ]
+ },
+ "MCQBlock1a2": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q11..20"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 476,
+ 370
+ ]
+ },
+ "MCQBlock1a3": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q21..35"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 476,
+ 638
+ ]
+ },
+ "MCQBlock2a1": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q51..60"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 645,
+ 100
+ ]
+ },
+ "MCQBlock2a2": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q61..70"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 645,
+ 370
+ ]
+ },
+ "MCQBlock2a3": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q71..85"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 645,
+ 638
+ ]
+ },
+ "MCQBlock3a1": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q101..110"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 815,
+ 100
+ ]
+ },
+ "MCQBlock3a2": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q111..120"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 815,
+ 370
+ ]
+ },
+ "MCQBlock3a3": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q121..135"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 815,
+ 638
+ ]
+ },
+ "MCQBlock4a1": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q151..160"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 983,
+ 100
+ ]
+ },
+ "MCQBlock4a2": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q161..170"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 983,
+ 370
+ ]
+ },
+ "MCQBlock4a3": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q171..185"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 983,
+ 638
+ ]
+ },
+ "MCQBlock1a": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q36..50"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 480,
+ 1061
+ ]
+ },
+ "MCQBlock2a": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q86..100"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 648,
+ 1061
+ ]
+ },
+ "MCQBlock3a": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q136..150"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 815,
+ 1061
+ ]
+ },
+ "MCQBlock4a": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q186..200"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.6,
+ "origin": [
+ 986,
+ 1061
+ ]
+ }
+ }
+}
diff --git a/samples/community/Shamanth/omr_sheet_01.png b/samples/community/Shamanth/omr_sheet_01.png
new file mode 100644
index 00000000..ead17dd1
Binary files /dev/null and b/samples/community/Shamanth/omr_sheet_01.png differ
diff --git a/samples/community/Shamanth/template.json b/samples/community/Shamanth/template.json
new file mode 100644
index 00000000..fb18d975
--- /dev/null
+++ b/samples/community/Shamanth/template.json
@@ -0,0 +1,25 @@
+{
+ "pageDimensions": [
+ 300,
+ 400
+ ],
+ "bubbleDimensions": [
+ 20,
+ 20
+ ],
+ "fieldBlocks": {
+ "MCQBlock1": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 78,
+ 41
+ ],
+ "fieldLabels": [
+ "q21..28"
+ ],
+ "bubblesGap": 56,
+ "labelsGap": 46
+ }
+ },
+ "preProcessors": []
+}
diff --git a/samples/community/UPSC-mock/answer_key.jpg b/samples/community/UPSC-mock/answer_key.jpg
new file mode 100644
index 00000000..7a229c1b
Binary files /dev/null and b/samples/community/UPSC-mock/answer_key.jpg differ
diff --git a/samples/community/UPSC-mock/config.json b/samples/community/UPSC-mock/config.json
new file mode 100644
index 00000000..0edf9d22
--- /dev/null
+++ b/samples/community/UPSC-mock/config.json
@@ -0,0 +1,11 @@
+{
+ "dimensions": {
+ "display_height": 1800,
+ "display_width": 2400,
+ "processing_height": 2400,
+ "processing_width": 1800
+ },
+ "outputs": {
+ "show_image_level": 0
+ }
+}
diff --git a/samples/community/UPSC-mock/evaluation.json b/samples/community/UPSC-mock/evaluation.json
new file mode 100644
index 00000000..42fbec8b
--- /dev/null
+++ b/samples/community/UPSC-mock/evaluation.json
@@ -0,0 +1,18 @@
+{
+ "source_type": "csv",
+ "options": {
+ "answer_key_csv_path": "answer_key.csv",
+ "answer_key_image_path": "answer_key.jpg",
+ "questions_in_order": [
+ "q1..100"
+ ],
+ "should_explain_scoring": true
+ },
+ "marking_schemes": {
+ "DEFAULT": {
+ "correct": "2",
+ "incorrect": "-2/3",
+ "unmarked": "0"
+ }
+ }
+}
diff --git a/samples/community/UPSC-mock/scan-angles/angle-1.jpg b/samples/community/UPSC-mock/scan-angles/angle-1.jpg
new file mode 100644
index 00000000..9c75d7be
Binary files /dev/null and b/samples/community/UPSC-mock/scan-angles/angle-1.jpg differ
diff --git a/samples/community/UPSC-mock/scan-angles/angle-2.jpg b/samples/community/UPSC-mock/scan-angles/angle-2.jpg
new file mode 100644
index 00000000..edab6060
Binary files /dev/null and b/samples/community/UPSC-mock/scan-angles/angle-2.jpg differ
diff --git a/samples/community/UPSC-mock/scan-angles/angle-3.jpg b/samples/community/UPSC-mock/scan-angles/angle-3.jpg
new file mode 100644
index 00000000..f1da4af9
Binary files /dev/null and b/samples/community/UPSC-mock/scan-angles/angle-3.jpg differ
diff --git a/samples/community/UPSC-mock/template.json b/samples/community/UPSC-mock/template.json
new file mode 100644
index 00000000..a1ef5c6b
--- /dev/null
+++ b/samples/community/UPSC-mock/template.json
@@ -0,0 +1,195 @@
+{
+ "pageDimensions": [
+ 1800,
+ 2400
+ ],
+ "bubbleDimensions": [
+ 30,
+ 25
+ ],
+ "customLabels": {
+ "Subject Code": [
+ "subjectCode1",
+ "subjectCode2"
+ ],
+ "Roll": [
+ "roll1..10"
+ ]
+ },
+ "fieldBlocks": {
+ "bookletNo": {
+ "origin": [
+ 595,
+ 545
+ ],
+ "bubblesGap": 68,
+ "labelsGap": 0,
+ "fieldLabels": [
+ "bookletNo"
+ ],
+ "bubbleValues": [
+ "A",
+ "B",
+ "C",
+ "D"
+ ],
+ "direction": "vertical"
+ },
+ "subjectCode": {
+ "origin": [
+ 912,
+ 512
+ ],
+ "bubblesGap": 33,
+ "labelsGap": 42.5,
+ "fieldLabels": [
+ "subjectCode1",
+ "subjectCode2"
+ ],
+ "fieldType": "QTYPE_INT"
+ },
+ "roll": {
+ "origin": [
+ 1200,
+ 510
+ ],
+ "bubblesGap": 33,
+ "labelsGap": 42.8,
+ "fieldLabels": [
+ "roll1..10"
+ ],
+ "fieldType": "QTYPE_INT"
+ },
+ "q01block": {
+ "origin": [
+ 500,
+ 927
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q1..10"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q11block": {
+ "origin": [
+ 500,
+ 1258
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q11..20"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q21block": {
+ "origin": [
+ 500,
+ 1589
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q21..30"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q31block": {
+ "origin": [
+ 495,
+ 1925
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q31..40"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q41block": {
+ "origin": [
+ 811,
+ 927
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q41..50"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q51block": {
+ "origin": [
+ 811,
+ 1258
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q51..60"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q61block": {
+ "origin": [
+ 811,
+ 1589
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q61..70"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q71block": {
+ "origin": [
+ 811,
+ 1925
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q71..80"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q81block": {
+ "origin": [
+ 1125,
+ 927
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q81..90"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q91block": {
+ "origin": [
+ 1125,
+ 1258
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q91..100"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ }
+ },
+ "preProcessors": [
+ {
+ "name": "CropPage",
+ "options": {
+ "morphKernel": [
+ 10,
+ 10
+ ]
+ }
+ }
+ ]
+}
diff --git a/samples/community/UmarFarootAPS/answer_key.csv b/samples/community/UmarFarootAPS/answer_key.csv
new file mode 100644
index 00000000..b40e8959
--- /dev/null
+++ b/samples/community/UmarFarootAPS/answer_key.csv
@@ -0,0 +1,200 @@
+q1,C
+q2,C
+q3,"D,E"
+q4,"A,AB"
+q5,"[['A', '1'], ['B', '2']]"
+q6,"['A', 'B']"
+q7,C
+q8,D
+q9,B
+q10,B
+q11,A
+q12,A
+q13,C
+q14,B
+q15,D
+q16,B
+q17,C
+q18,A
+q19,B
+q20,D
+q21,D
+q22,C
+q23,A
+q24,C
+q25,D
+q26,C
+q27,C
+q28,B
+q29,A
+q30,D
+q31,C
+q32,B
+q33,B
+q34,C
+q35,A
+q36,D
+q37,C
+q38,B
+q39,C
+q40,A
+q41,A
+q42,C
+q43,D
+q44,D
+q45,B
+q46,C
+q47,C
+q48,A
+q49,C
+q50,B
+q51,B
+q52,C
+q53,D
+q54,C
+q55,B
+q56,B
+q57,A
+q58,A
+q59,D
+q60,C
+q61,C
+q62,B
+q63,A
+q64,C
+q65,D
+q66,C
+q67,B
+q68,A
+q69,B
+q70,B
+q71,C
+q72,B
+q73,C
+q74,A
+q75,A
+q76,C
+q77,D
+q78,D
+q79,B
+q80,A
+q81,B
+q82,C
+q83,D
+q84,C
+q85,A
+q86,C
+q87,D
+q88,B
+q89,C
+q90,B
+q91,B
+q92,A
+q93,C
+q94,D
+q95,C
+q96,B
+q97,B
+q98,A
+q99,A
+q100,A
+q101,A
+q102,B
+q103,C
+q104,C
+q105,A
+q106,D
+q107,B
+q108,A
+q109,C
+q110,B
+q111,B
+q112,C
+q113,C
+q114,B
+q115,D
+q116,B
+q117,A
+q118,C
+q119,D
+q120,C
+q121,C
+q122,A
+q123,B
+q124,C
+q125,D
+q126,C
+q127,C
+q128,D
+q129,D
+q130,A
+q131,A
+q132,C
+q133,B
+q134,C
+q135,D
+q136,B
+q137,C
+q138,A
+q139,B
+q140,D
+q141,D
+q142,C
+q143,D
+q144,A
+q145,A
+q146,C
+q147,A
+q148,D
+q149,D
+q150,B
+q151,A
+q152,B
+q153,B
+q154,D
+q155,D
+q156,B
+q157,A
+q158,B
+q159,A
+q160,C
+q161,D
+q162,C
+q163,A
+q164,B
+q165,D
+q166,D
+q167,C
+q168,C
+q169,C
+q170,D
+q171,A
+q172,A
+q173,C
+q174,C
+q175,B
+q176,D
+q177,A
+q178,B
+q179,B
+q180,C
+q181,D
+q182,C
+q183,B
+q184,B
+q185,C
+q186,D
+q187,D
+q188,A
+q189,A
+q190,B
+q191,C
+q192,B
+q193,D
+q194,C
+q195,B
+q196,B
+q197,A
+q198,B
+q199,B
+q200,A
diff --git a/samples/community/UmarFarootAPS/config.json b/samples/community/UmarFarootAPS/config.json
new file mode 100644
index 00000000..fde8f5b2
--- /dev/null
+++ b/samples/community/UmarFarootAPS/config.json
@@ -0,0 +1,11 @@
+{
+ "dimensions": {
+ "display_height": 960,
+ "display_width": 1280,
+ "processing_height": 1640,
+ "processing_width": 1332
+ },
+ "outputs": {
+ "show_image_level": 0
+ }
+}
diff --git a/samples/community/UmarFarootAPS/evaluation.json b/samples/community/UmarFarootAPS/evaluation.json
new file mode 100644
index 00000000..14a3db25
--- /dev/null
+++ b/samples/community/UmarFarootAPS/evaluation.json
@@ -0,0 +1,14 @@
+{
+ "source_type": "csv",
+ "options": {
+ "answer_key_csv_path": "answer_key.csv",
+ "should_explain_scoring": true
+ },
+ "marking_schemes": {
+ "DEFAULT": {
+ "correct": "1",
+ "incorrect": "0",
+ "unmarked": "0"
+ }
+ }
+}
diff --git a/samples/sample3/omr_marker.jpg b/samples/community/UmarFarootAPS/omr_marker.jpg
similarity index 100%
rename from samples/sample3/omr_marker.jpg
rename to samples/community/UmarFarootAPS/omr_marker.jpg
diff --git a/samples/community/UmarFarootAPS/scans/scan-type-1.jpg b/samples/community/UmarFarootAPS/scans/scan-type-1.jpg
new file mode 100644
index 00000000..0932b240
Binary files /dev/null and b/samples/community/UmarFarootAPS/scans/scan-type-1.jpg differ
diff --git a/samples/community/UmarFarootAPS/scans/scan-type-2.jpg b/samples/community/UmarFarootAPS/scans/scan-type-2.jpg
new file mode 100644
index 00000000..2eef7c32
Binary files /dev/null and b/samples/community/UmarFarootAPS/scans/scan-type-2.jpg differ
diff --git a/samples/community/UmarFarootAPS/template.json b/samples/community/UmarFarootAPS/template.json
new file mode 100644
index 00000000..f096f518
--- /dev/null
+++ b/samples/community/UmarFarootAPS/template.json
@@ -0,0 +1,188 @@
+{
+ "pageDimensions": [
+ 2550,
+ 3300
+ ],
+ "bubbleDimensions": [
+ 32,
+ 32
+ ],
+ "preProcessors": [
+ {
+ "name": "CropOnMarkers",
+ "options": {
+ "relativePath": "omr_marker.jpg",
+ "sheetToMarkerWidthRatio": 17
+ }
+ }
+ ],
+ "customLabels": {
+ "Roll_no": [
+ "r1",
+ "r2",
+ "r3",
+ "r4"
+ ]
+ },
+ "fieldBlocks": {
+ "Roll_no": {
+ "fieldType": "QTYPE_INT",
+ "origin": [
+ 2169,
+ 180
+ ],
+ "fieldLabels": [
+ "r1",
+ "r2",
+ "r3",
+ "r4"
+ ],
+ "bubblesGap": 61,
+ "labelsGap": 93
+ },
+ "MCQBlock1a1": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 197,
+ 300
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q1..17"
+ ]
+ },
+ "MCQBlock1a2": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 197,
+ 1310
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q18..34"
+ ]
+ },
+ "MCQBlock1a3": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 197,
+ 2316
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q35..50"
+ ]
+ },
+ "MCQBlock1a4": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 725,
+ 300
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q51..67"
+ ]
+ },
+ "MCQBlock1a5": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 725,
+ 1310
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q68..84"
+ ]
+ },
+ "MCQBlock1a6": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 725,
+ 2316
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q85..100"
+ ]
+ },
+ "MCQBlock1a7": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 1250,
+ 300
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q101..117"
+ ]
+ },
+ "MCQBlock1a8": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 1250,
+ 1310
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q118..134"
+ ]
+ },
+ "MCQBlock1a9": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 1250,
+ 2316
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q135..150"
+ ]
+ },
+ "MCQBlock1a10": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 1770,
+ 300
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q151..167"
+ ]
+ },
+ "MCQBlock1a11": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 1770,
+ 1310
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q168..184"
+ ]
+ },
+ "MCQBlock1a12": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 1770,
+ 2316
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q185..200"
+ ]
+ }
+ }
+}
diff --git a/samples/community/dxuian/omrcollegesheet.jpg b/samples/community/dxuian/omrcollegesheet.jpg
new file mode 100644
index 00000000..b9d70c6b
Binary files /dev/null and b/samples/community/dxuian/omrcollegesheet.jpg differ
diff --git a/samples/community/dxuian/template.json b/samples/community/dxuian/template.json
new file mode 100644
index 00000000..43bf2be6
--- /dev/null
+++ b/samples/community/dxuian/template.json
@@ -0,0 +1,48 @@
+{
+ "pageDimensions": [707, 484],
+ "bubbleDimensions": [15, 10],
+ "fieldBlocks": {
+ "Column1": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [82, 35],
+ "bubblesGap": 21,
+ "labelsGap": 22.7,
+ "bubbleCount": 20,
+ "fieldLabels": ["Q1", "Q2", "Q3", "Q4", "Q5", "Q6", "Q7", "Q8", "Q9", "Q10", "Q11", "Q12", "Q13", "Q14", "Q15", "Q16", "Q17", "Q18", "Q19", "Q20"]
+ },
+ "Column2": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [205, 35],
+ "bubblesGap": 21,
+ "labelsGap": 22.7,
+ "bubbleCount": 20,
+ "fieldLabels": ["Q21", "Q22", "Q23", "Q24", "Q25", "Q26", "Q27", "Q28", "Q29", "Q30", "Q31", "Q32", "Q33", "Q34", "Q35", "Q36", "Q37", "Q38", "Q39", "Q40"]
+ },
+ "Column3": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [327, 35],
+ "bubblesGap": 21,
+ "labelsGap": 22.7,
+ "bubbleCount": 20,
+ "fieldLabels": ["Q41", "Q42", "Q43", "Q44", "Q45", "Q46", "Q47", "Q48", "Q49", "Q50", "Q51", "Q52", "Q53", "Q54", "Q55", "Q56", "Q57", "Q58", "Q59", "Q60"]
+ },
+ "Column4": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [450, 35],
+ "bubblesGap": 21,
+ "labelsGap": 22.7,
+ "bubbleCount": 20,
+ "fieldLabels": ["Q61", "Q62", "Q63", "Q64", "Q65", "Q66", "Q67", "Q68", "Q69", "Q70", "Q71", "Q72", "Q73", "Q74", "Q75", "Q76", "Q77", "Q78", "Q79", "Q80"]
+ },
+ "Column5": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [573, 35],
+ "bubblesGap": 21,
+ "labelsGap": 22.7,
+ "bubbleCount": 20,
+ "fieldLabels": ["Q81", "Q82", "Q83", "Q84", "Q85", "Q86", "Q87", "Q88", "Q89", "Q90", "Q91", "Q92", "Q93", "Q94", "Q95", "Q96", "Q97", "Q98", "Q99", "Q100"]
+ }
+ },
+
+ "emptyValue": "-"
+}
\ No newline at end of file
diff --git a/samples/community/ibrahimkilic/template.json b/samples/community/ibrahimkilic/template.json
new file mode 100644
index 00000000..f0cb9cf3
--- /dev/null
+++ b/samples/community/ibrahimkilic/template.json
@@ -0,0 +1,30 @@
+{
+ "pageDimensions": [
+ 299,
+ 328
+ ],
+ "bubbleDimensions": [
+ 20,
+ 20
+ ],
+ "emptyValue": "no",
+ "fieldBlocks": {
+ "YesNoBlock1": {
+ "direction": "horizontal",
+ "bubbleValues": [
+ "yes"
+ ],
+ "origin": [
+ 15,
+ 55
+ ],
+ "emptyValue": "no",
+ "bubblesGap": 48,
+ "labelsGap": 48,
+ "fieldLabels": [
+ "q1..5"
+ ]
+ }
+ },
+ "preProcessors": []
+}
diff --git a/samples/community/ibrahimkilic/yes_no_questionnarie.jpg b/samples/community/ibrahimkilic/yes_no_questionnarie.jpg
new file mode 100644
index 00000000..e9436f41
Binary files /dev/null and b/samples/community/ibrahimkilic/yes_no_questionnarie.jpg differ
diff --git a/samples/community/samuelIkoli/template.json b/samples/community/samuelIkoli/template.json
new file mode 100644
index 00000000..759f6972
--- /dev/null
+++ b/samples/community/samuelIkoli/template.json
@@ -0,0 +1,28 @@
+{
+ "pageDimensions": [630, 404],
+ "bubbleDimensions": [20, 15],
+ "customLabels": {},
+ "fieldBlocks": {
+ "MCQBlock1": {
+ "fieldType": "QTYPE_MCQ5",
+ "origin": [33, 6],
+ "fieldLabels": ["q1", "q2", "q3", "q4", "q5", "q6", "q7", "q8", "q9", "q10", "q11", "q12", "q13", "q14", "q15", "q16", "q17", "q18", "q19", "q20"],
+ "bubblesGap": 33,
+ "labelsGap": 20
+ },
+ "MCQBlock2": {
+ "fieldType": "QTYPE_MCQ5",
+ "origin": [248, 6],
+ "fieldLabels": ["q21", "q22", "q23", "q24", "q25", "q26", "q27", "q28", "q29", "q30", "q31", "q32", "q33", "q34", "q35", "q36", "q37", "q38", "q39", "q40"],
+ "bubblesGap": 33,
+ "labelsGap": 20
+ },
+ "MCQBlock3": {
+ "fieldType": "QTYPE_MCQ5",
+ "origin": [465, 6],
+ "fieldLabels": ["q41", "q42", "q43", "q44", "q45", "q46", "q47", "q48", "q49", "q50", "q51", "q52", "q53", "q54", "q55", "q56", "q57", "q58", "q59", "q60"],
+ "bubblesGap": 33,
+ "labelsGap": 20
+ }
+ }
+}
diff --git a/samples/community/samuelIkoli/waec_sample.jpeg b/samples/community/samuelIkoli/waec_sample.jpeg
new file mode 100644
index 00000000..fcd61e80
Binary files /dev/null and b/samples/community/samuelIkoli/waec_sample.jpeg differ
diff --git a/samples/sample1/config.json b/samples/sample1/config.json
new file mode 100644
index 00000000..a9daf449
--- /dev/null
+++ b/samples/sample1/config.json
@@ -0,0 +1,11 @@
+{
+ "dimensions": {
+ "display_height": 2480,
+ "display_width": 1640,
+ "processing_height": 820,
+ "processing_width": 666
+ },
+ "outputs": {
+ "show_image_level": 5
+ }
+}
diff --git a/samples/sample1/template.json b/samples/sample1/template.json
index ec0f9c68..46635774 100644
--- a/samples/sample1/template.json
+++ b/samples/sample1/template.json
@@ -1,260 +1,197 @@
{
- "Dimensions": [
+ "pageDimensions": [
1846,
1500
],
- "BubbleDimensions": [
+ "bubbleDimensions": [
40,
40
],
- "Options": {
- "Marker": {
- "RelativePath": "omr_marker.jpg",
- "SheetToMarkerWidthRatio": 17
- }
- },
- "Concatenations": {
+ "customLabels": {
"Roll": [
- "Squad",
"Medium",
- "roll0",
- "roll1",
- "roll2",
- "roll3",
- "roll4",
- "roll5",
- "roll6",
- "roll7",
- "roll8"
+ "roll1..9"
],
"q5": [
- "q5.1",
- "q5.2"
+ "q5_1",
+ "q5_2"
],
"q6": [
- "q6.1",
- "q6.2"
+ "q6_1",
+ "q6_2"
],
"q7": [
- "q7.1",
- "q7.2"
+ "q7_1",
+ "q7_2"
],
- "q9": [
- "q9.1",
- "q9.2"
+ "q8": [
+ "q8_1",
+ "q8_2"
],
- "q11": [
- "q8.1",
- "q8.2"
+ "q9": [
+ "q9_1",
+ "q9_2"
]
},
- "Singles": [
- "q1",
- "q2",
- "q3",
- "q4",
- "q10",
- "q11",
- "q12",
- "q13",
- "q14",
- "q15",
- "q16",
- "q17",
- "q18",
- "q19",
- "q20"
- ],
- "QBlocks": {
+ "fieldBlocks": {
"Medium": {
- "qType": "QTYPE_MED",
- "orig": [
- 160,
- 285
- ],
- "bigGaps": [
- 115,
- 11
- ],
- "gaps": [
- 59,
- 46
- ],
- "qNos": [
- [
- [
- "Medium"
- ]
- ]
+ "bubblesGap": 41,
+ "bubbleValues": [
+ "E",
+ "H"
+ ],
+ "direction": "vertical",
+ "fieldLabels": [
+ "Medium"
+ ],
+ "labelsGap": 0,
+ "origin": [
+ 170,
+ 282
]
},
"Roll": {
- "qType": "QTYPE_ROLL",
- "orig": [
- 218,
- 285
- ],
- "bigGaps": [
- 115,
- 11
- ],
- "gaps": [
- 58,
- 46
- ],
- "qNos": [
- [
- [
- "roll0",
- "roll1",
- "roll2",
- "roll3",
- "roll4",
- "roll5",
- "roll6",
- "roll7",
- "roll8"
- ]
- ]
+ "fieldType": "QTYPE_INT",
+ "fieldLabels": [
+ "roll1..9"
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 58,
+ "origin": [
+ 225,
+ 282
]
},
- "Int1": {
- "qType": "QTYPE_INT",
- "orig": [
+ "Int_Block_Q5": {
+ "fieldType": "QTYPE_INT",
+ "fieldLabels": [
+ "q5_1",
+ "q5_2"
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 60,
+ "origin": [
903,
- 285
- ],
- "bigGaps": [
- 128,
- 11
- ],
- "gaps": [
- 59,
- 46
- ],
- "qNos": [
- [
- [
- "q5.1",
- "q5.2"
- ],
- [
- "q6.1",
- "q6.2"
- ],
- [
- "q7.1",
- "q7.2"
- ]
- ]
+ 282
]
},
- "Int2": {
- "qType": "QTYPE_INT",
- "orig": [
- 1418,
- 285
- ],
- "bigGaps": [
- 128,
- 11
- ],
- "gaps": [
- 59,
- 46
- ],
- "qNos": [
- [
- [
- "q8.1",
- "q8.2"
- ],
- [
- "q9.1",
- "q9.2"
- ]
- ]
+ "Int_Block_Q6": {
+ "fieldType": "QTYPE_INT",
+ "fieldLabels": [
+ "q6_1",
+ "q6_2"
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 60,
+ "origin": [
+ 1077,
+ 282
]
},
- "Mcq1": {
- "qType": "QTYPE_MCQ4",
- "orig": [
- 118,
- 857
- ],
- "bigGaps": [
- 115,
- 181
- ],
- "gaps": [
- 59,
- 53
- ],
- "qNos": [
- [
- [
- "q1",
- "q2",
- "q3",
- "q4"
- ],
- [
- "q10",
- "q11",
- "q12",
- "q13"
- ]
- ]
+ "Int_Block_Q7": {
+ "fieldType": "QTYPE_INT",
+ "fieldLabels": [
+ "q7_1",
+ "q7_2"
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 60,
+ "origin": [
+ 1240,
+ 282
+ ]
+ },
+ "Int_Block_Q8": {
+ "fieldType": "QTYPE_INT",
+ "fieldLabels": [
+ "q8_1",
+ "q8_2"
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 57,
+ "origin": [
+ 1410,
+ 282
+ ]
+ },
+ "Int_Block_Q9": {
+ "fieldType": "QTYPE_INT",
+ "fieldLabels": [
+ "q9_1",
+ "q9_2"
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 57,
+ "origin": [
+ 1580,
+ 282
]
},
- "Mcq2": {
- "qType": "QTYPE_MCQ4",
- "orig": [
+ "MCQ_Block_Q1": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q1..4"
+ ],
+ "bubblesGap": 59,
+ "labelsGap": 50,
+ "origin": [
+ 121,
+ 860
+ ]
+ },
+ "MCQ_Block_Q10": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q10..13"
+ ],
+ "bubblesGap": 59,
+ "labelsGap": 50,
+ "origin": [
+ 121,
+ 1195
+ ]
+ },
+ "MCQ_Block_Q14": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q14..16"
+ ],
+ "bubblesGap": 57,
+ "labelsGap": 50,
+ "origin": [
905,
860
- ],
- "bigGaps": [
- 115,
- 180
- ],
- "gaps": [
- 59,
- 53
- ],
- "qNos": [
- [
- [
- "q14",
- "q15",
- "q16"
- ]
- ]
]
},
- "Mcq3": {
- "qType": "QTYPE_MCQ4",
- "orig": [
+ "MCQ_Block_Q17": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q17..20"
+ ],
+ "bubblesGap": 57,
+ "labelsGap": 50,
+ "origin": [
905,
- 1198
- ],
- "bigGaps": [
- 115,
- 180
- ],
- "gaps": [
- 59,
- 53
- ],
- "qNos": [
- [
- [
- "q17",
- "q18",
- "q19",
- "q20"
- ]
- ]
+ 1195
]
}
- }
-}
\ No newline at end of file
+ },
+ "preProcessors": [
+ {
+ "name": "CropPage",
+ "options": {
+ "morphKernel": [
+ 10,
+ 10
+ ]
+ }
+ },
+ {
+ "name": "CropOnMarkers",
+ "options": {
+ "relativePath": "omr_marker.jpg",
+ "sheetToMarkerWidthRatio": 17
+ }
+ }
+ ]
+}
diff --git a/samples/sample2/AdrianSample/adrian_omr.png b/samples/sample2/AdrianSample/adrian_omr.png
index dc008145..69a53823 100644
Binary files a/samples/sample2/AdrianSample/adrian_omr.png and b/samples/sample2/AdrianSample/adrian_omr.png differ
diff --git a/samples/sample2/AdrianSample/adrian_omr_2.png b/samples/sample2/AdrianSample/adrian_omr_2.png
new file mode 100644
index 00000000..d8db0994
Binary files /dev/null and b/samples/sample2/AdrianSample/adrian_omr_2.png differ
diff --git a/samples/sample2/config.json b/samples/sample2/config.json
new file mode 100644
index 00000000..a9daf449
--- /dev/null
+++ b/samples/sample2/config.json
@@ -0,0 +1,11 @@
+{
+ "dimensions": {
+ "display_height": 2480,
+ "display_width": 1640,
+ "processing_height": 820,
+ "processing_width": 666
+ },
+ "outputs": {
+ "show_image_level": 5
+ }
+}
diff --git a/samples/sample2/template.json b/samples/sample2/template.json
index e82a9051..41ec9ffa 100644
--- a/samples/sample2/template.json
+++ b/samples/sample2/template.json
@@ -1,46 +1,35 @@
{
- "Dimensions": [
+ "pageDimensions": [
300,
400
],
- "BubbleDimensions": [
+ "bubbleDimensions": [
25,
25
],
- "Concatenations": {},
- "Singles": [
- "q1",
- "q2",
- "q3",
- "q4",
- "q5"
+ "preProcessors": [
+ {
+ "name": "CropPage",
+ "options": {
+ "morphKernel": [
+ 10,
+ 10
+ ]
+ }
+ }
],
- "QBlocks": {
- "MCQBlock1": {
- "qType": "QTYPE_MCQ5",
- "orig": [
+ "fieldBlocks": {
+ "MCQ_Block_1": {
+ "fieldType": "QTYPE_MCQ5",
+ "origin": [
65,
60
],
- "qNos": [
- [
- [
- "q1",
- "q2",
- "q3",
- "q4",
- "q5"
- ]
- ]
- ],
- "gaps": [
- 41,
- 52
+ "fieldLabels": [
+ "q1..5"
],
- "bigGaps": [
- 30,
- 30
- ]
+ "labelsGap": 52,
+ "bubblesGap": 41
}
}
-}
\ No newline at end of file
+}
diff --git a/samples/sample3/README.md b/samples/sample3/README.md
new file mode 100644
index 00000000..d764c8b3
--- /dev/null
+++ b/samples/sample3/README.md
@@ -0,0 +1,31 @@
+## Observation
+The OMR layout is slightly different on colored thick papers vs on xeroxed thin papers. The shift becomes noticible in case of OMR with large number of questions.
+
+We overlapped a colored OMR sheet with a xerox copy of the same OMR sheet(both printed on A4 papers) and observed that there is a great amount of layout sheet as we reach the bottom of the OMR.
+
+Link to an explainer with a real life example: [Google Drive](https://drive.google.com/drive/folders/1GpZTmpEhEjSALJEMjHwDafzWKgEoCTOI?usp=sharing)
+
+## Reasons for shifts in Template layout:
+Listing out a few reasons for the above observation:
+### Printer margin setting
+The margin settings for different printers may be different for the same OMR layout. Thus causing the print to become elongated either horizontally, vertically or both ways.
+
+### The Fan-out effect
+The fan-out effect is usually observed in a sheet fed offset press. Depending on how the papers are made, their dimensions have a tendency to change when they are exposed to moisture or water.
+
+The standard office papers(80 gsm) can easily capture moisture and change shape e.g. in case they get stored for multiple days in a place where the weather is highly humid.
+
+Below are some examples of the GSM ranges:
+
+- 74gsm to 90gsm – This is the basic standard office paper, used in your laser printers.
+- 100gsm to 120gsm – This is stationary paper used for standard letterheads, complimentary slips.
+- 130 to 170gsm – Mostly used for leaflets, posters, single-sided flyers, and brochures.
+
+## Solution
+
+It is recommended to scan each types of prints into different folders and use a separate template.json layout for each of the folders. The same is presented in this sample folder.
+
+## References
+
+- [Paper dimensional stability in sheet-fed offset printing](https://www.diva-portal.org/smash/get/diva2:517895/FULLTEXT01.pdf)
+- An analysis of a few ["Interesting" bubble sheets](https://imgur.com/a/10qwL)
diff --git a/samples/sample3/colored-thick-sheet/rgb-100-gsm.jpg b/samples/sample3/colored-thick-sheet/rgb-100-gsm.jpg
new file mode 100644
index 00000000..4a34731a
Binary files /dev/null and b/samples/sample3/colored-thick-sheet/rgb-100-gsm.jpg differ
diff --git a/samples/sample3/colored-thick-sheet/template.json b/samples/sample3/colored-thick-sheet/template.json
new file mode 100644
index 00000000..743723d6
--- /dev/null
+++ b/samples/sample3/colored-thick-sheet/template.json
@@ -0,0 +1,143 @@
+{
+ "pageDimensions": [
+ 1800,
+ 2400
+ ],
+ "bubbleDimensions": [
+ 23,
+ 20
+ ],
+ "fieldBlocks": {
+ "q01block": {
+ "origin": [
+ 504,
+ 927
+ ],
+ "bubblesGap": 60.35,
+ "labelsGap": 31.75,
+ "fieldLabels": [
+ "q1..10"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q11block": {
+ "origin": [
+ 504,
+ 1242
+ ],
+ "bubblesGap": 60.35,
+ "labelsGap": 31.75,
+ "fieldLabels": [
+ "q11..20"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q21block": {
+ "origin": [
+ 500,
+ 1562
+ ],
+ "bubblesGap": 61.25,
+ "labelsGap": 32.5,
+ "fieldLabels": [
+ "q21..30"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q31block": {
+ "origin": [
+ 500,
+ 1885
+ ],
+ "bubblesGap": 62.25,
+ "labelsGap": 33.5,
+ "fieldLabels": [
+ "q31..40"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q41block": {
+ "origin": [
+ 811,
+ 927
+ ],
+ "bubblesGap": 60.35,
+ "labelsGap": 31.75,
+ "fieldLabels": [
+ "q41..50"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q51block": {
+ "origin": [
+ 811,
+ 1242
+ ],
+ "bubblesGap": 60.35,
+ "labelsGap": 31.75,
+ "fieldLabels": [
+ "q51..60"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q61block": {
+ "origin": [
+ 811,
+ 1562
+ ],
+ "bubblesGap": 61.25,
+ "labelsGap": 32.5,
+ "fieldLabels": [
+ "q61..70"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q71block": {
+ "origin": [
+ 811,
+ 1885
+ ],
+ "bubblesGap": 62.25,
+ "labelsGap": 33.5,
+ "fieldLabels": [
+ "q71..80"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q81block": {
+ "origin": [
+ 1120,
+ 927
+ ],
+ "bubblesGap": 60.35,
+ "labelsGap": 31.75,
+ "fieldLabels": [
+ "q81..90"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q91block": {
+ "origin": [
+ 1120,
+ 1242
+ ],
+ "bubblesGap": 60.35,
+ "labelsGap": 31.75,
+ "fieldLabels": [
+ "q91..100"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ }
+ },
+ "preProcessors": [
+ {
+ "name": "CropPage",
+ "options": {
+ "morphKernel": [
+ 10,
+ 10
+ ]
+ }
+ }
+ ]
+}
diff --git a/samples/sample3/config.json b/samples/sample3/config.json
new file mode 100644
index 00000000..a9daf449
--- /dev/null
+++ b/samples/sample3/config.json
@@ -0,0 +1,11 @@
+{
+ "dimensions": {
+ "display_height": 2480,
+ "display_width": 1640,
+ "processing_height": 820,
+ "processing_width": 666
+ },
+ "outputs": {
+ "show_image_level": 5
+ }
+}
diff --git a/samples/sample3/template.json b/samples/sample3/template.json
deleted file mode 100644
index 2dddf37e..00000000
--- a/samples/sample3/template.json
+++ /dev/null
@@ -1,248 +0,0 @@
-{
- "Dimensions": [
- 1846,
- 1500
- ],
- "BubbleDimensions": [
- 40,
- 40
- ],
- "Options": {
- "Marker": {
- "RelativePath": "omr_marker.jpg",
- "SheetToMarkerWidthRatio": 17
- },
- "OverrideFlags":{
- "noCropping": true
- }
- },
- "Concatenations": {
- "Roll": [
- "Squad",
- "Medium",
- "roll0",
- "roll1",
- "roll2",
- "roll3",
- "roll4",
- "roll5",
- "roll6",
- "roll7",
- "roll8"
- ],
- "q6": [
- "q6.1",
- "q6.2"
- ],
- "q7": [
- "q7.1",
- "q7.2"
- ],
- "q8": [
- "q8.1",
- "q8.2"
- ],
- "q9": [
- "q9.1",
- "q9.2"
- ],
- "q10": [
- "q10.1",
- "q10.2"
- ]
- },
- "Singles": [
- "q1",
- "q2",
- "q3",
- "q4",
- "q5",
- "q11",
- "q12",
- "q13",
- "q14",
- "q15",
- "q16",
- "q17",
- "q18",
- "q19",
- "q20",
- "q21",
- "q22"
- ],
- "QBlocks": {
- "Medium": {
- "qType": "QTYPE_MED",
- "orig": [
- 208,
- 205
- ],
- "bigGaps": [
- 115,
- 11
- ],
- "gaps": [
- 59,
- 46
- ],
- "qNos": [
- [
- [
- "Medium"
- ]
- ]
- ]
- },
- "Roll": {
- "qType": "QTYPE_ROLL",
- "orig": [
- 261,
- 210
- ],
- "bigGaps": [
- 115,
- 11
- ],
- "gaps": [
- 58,
- 46
- ],
- "qNos": [
- [
- [
- "roll0",
- "roll1",
- "roll2",
- "roll3",
- "roll4",
- "roll5",
- "roll6",
- "roll7",
- "roll8"
- ]
- ]
- ]
- },
- "Int1": {
- "qType": "QTYPE_INT",
- "orig": [
- 935,
- 211
- ],
- "bigGaps": [
- 124,
- 11
- ],
- "gaps": [
- 57,
- 46
- ],
- "qNos": [
- [
- [
- "q6.1",
- "q6.2"
- ],
- [
- "q7.1",
- "q7.2"
- ],
- [
- "q8.1",
- "q8.2"
- ],
- [
- "q9.1",
- "q9.2"
- ],
- [
- "q10.1",
- "q10.2"
- ]
- ]
- ]
- },
- "Mcq1": {
- "qType": "QTYPE_MCQ4",
- "orig": [
- 198,
- 826
- ],
- "bigGaps": [
- 115,
- 183
- ],
- "gaps": [
- 93,
- 62
- ],
- "qNos": [
- [
- [
- "q1",
- "q2",
- "q3",
- "q4",
- "q5"
- ]
- ]
- ]
- },
- "Mcq2": {
- "qType": "QTYPE_MCQ4",
- "orig": [
- 833,
- 830
- ],
- "bigGaps": [
- 127,
- 254
- ],
- "gaps": [
- 71,
- 61
- ],
- "qNos": [
- [
- [
- "q11",
- "q12",
- "q13",
- "q14"
- ],
- [
- "q15",
- "q16",
- "q17",
- "q18"
- ]
- ]
- ]
- },
- "Mcq3": {
- "qType": "QTYPE_MCQ4",
- "orig": [
- 1481,
- 830
- ],
- "bigGaps": [
- 115,
- 183
- ],
- "gaps": [
- 73,
- 61
- ],
- "qNos": [
- [
- [
- "q19",
- "q20",
- "q21",
- "q22"
- ]
- ]
- ]
- }
- }
-}
\ No newline at end of file
diff --git a/samples/sample3/xeroxed-thin-sheet/grayscale-80-gsm.jpg b/samples/sample3/xeroxed-thin-sheet/grayscale-80-gsm.jpg
new file mode 100644
index 00000000..3e0e3efa
Binary files /dev/null and b/samples/sample3/xeroxed-thin-sheet/grayscale-80-gsm.jpg differ
diff --git a/samples/sample3/xeroxed-thin-sheet/template.json b/samples/sample3/xeroxed-thin-sheet/template.json
new file mode 100644
index 00000000..8aa4e41c
--- /dev/null
+++ b/samples/sample3/xeroxed-thin-sheet/template.json
@@ -0,0 +1,143 @@
+{
+ "pageDimensions": [
+ 1800,
+ 2400
+ ],
+ "bubbleDimensions": [
+ 23,
+ 20
+ ],
+ "fieldBlocks": {
+ "q01block": {
+ "origin": [
+ 492,
+ 924
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q1..10"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q11block": {
+ "origin": [
+ 492,
+ 1258
+ ],
+ "bubblesGap": 59.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q11..20"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q21block": {
+ "origin": [
+ 492,
+ 1589
+ ],
+ "bubblesGap": 60.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q21..30"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q31block": {
+ "origin": [
+ 487,
+ 1920
+ ],
+ "bubblesGap": 61.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q31..40"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q41block": {
+ "origin": [
+ 807,
+ 924
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q41..50"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q51block": {
+ "origin": [
+ 803,
+ 1258
+ ],
+ "bubblesGap": 59.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q51..60"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q61block": {
+ "origin": [
+ 803,
+ 1589
+ ],
+ "bubblesGap": 60.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q61..70"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q71block": {
+ "origin": [
+ 803,
+ 1920
+ ],
+ "bubblesGap": 60.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q71..80"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q81block": {
+ "origin": [
+ 1115,
+ 924
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q81..90"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q91block": {
+ "origin": [
+ 1115,
+ 1258
+ ],
+ "bubblesGap": 59.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q91..100"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ }
+ },
+ "preProcessors": [
+ {
+ "name": "CropPage",
+ "options": {
+ "morphKernel": [
+ 10,
+ 10
+ ]
+ }
+ }
+ ]
+}
diff --git a/samples/sample4/Class1/sheet1.jpg b/samples/sample4/Class1/sheet1.jpg
deleted file mode 100644
index f3248903..00000000
Binary files a/samples/sample4/Class1/sheet1.jpg and /dev/null differ
diff --git a/samples/sample4/Class2/sheet1.jpg b/samples/sample4/Class2/sheet1.jpg
deleted file mode 100644
index f3248903..00000000
Binary files a/samples/sample4/Class2/sheet1.jpg and /dev/null differ
diff --git a/samples/sample4/IMG_20201116_143512.jpg b/samples/sample4/IMG_20201116_143512.jpg
new file mode 100644
index 00000000..2f591c7a
Binary files /dev/null and b/samples/sample4/IMG_20201116_143512.jpg differ
diff --git a/samples/sample4/IMG_20201116_150717658.jpg b/samples/sample4/IMG_20201116_150717658.jpg
new file mode 100644
index 00000000..592fe425
Binary files /dev/null and b/samples/sample4/IMG_20201116_150717658.jpg differ
diff --git a/samples/sample4/IMG_20201116_150750830.jpg b/samples/sample4/IMG_20201116_150750830.jpg
new file mode 100644
index 00000000..6846ef31
Binary files /dev/null and b/samples/sample4/IMG_20201116_150750830.jpg differ
diff --git a/samples/sample4/config.json b/samples/sample4/config.json
new file mode 100644
index 00000000..df59a460
--- /dev/null
+++ b/samples/sample4/config.json
@@ -0,0 +1,13 @@
+{
+ "dimensions": {
+ "display_width": 1189,
+ "display_height": 1682
+ },
+ "threshold_params": {
+ "MIN_JUMP": 30
+ },
+ "outputs": {
+ "filter_out_multimarked_files": false,
+ "show_image_level": 5
+ }
+}
diff --git a/samples/sample4/evaluation.json b/samples/sample4/evaluation.json
new file mode 100644
index 00000000..50db0fbe
--- /dev/null
+++ b/samples/sample4/evaluation.json
@@ -0,0 +1,34 @@
+{
+ "source_type": "custom",
+ "options": {
+ "questions_in_order": [
+ "q1..11"
+ ],
+ "answers_in_order": [
+ "B",
+ "D",
+ "C",
+ "B",
+ "D",
+ "C",
+ [
+ "B",
+ "C",
+ "BC"
+ ],
+ "A",
+ "C",
+ "D",
+ "C"
+ ],
+ "should_explain_scoring": true,
+ "enable_evaluation_table_to_csv": true
+ },
+ "marking_schemes": {
+ "DEFAULT": {
+ "correct": "3",
+ "incorrect": "-1",
+ "unmarked": "0"
+ }
+ }
+}
diff --git a/samples/sample4/omr_marker.jpg b/samples/sample4/omr_marker.jpg
deleted file mode 100644
index 201dab0a..00000000
Binary files a/samples/sample4/omr_marker.jpg and /dev/null differ
diff --git a/samples/sample4/template.json b/samples/sample4/template.json
index 958d6ef8..0d615b87 100644
--- a/samples/sample4/template.json
+++ b/samples/sample4/template.json
@@ -1,263 +1,45 @@
{
- "Dimensions": [
- 1846,
- 1500
+ "pageDimensions": [
+ 1189,
+ 1682
],
- "BubbleDimensions": [
- 40,
- 40
+ "bubbleDimensions": [
+ 30,
+ 30
],
- "Options": {
- "Marker": {
- "RelativePath": "omr_marker.jpg",
- "SheetToMarkerWidthRatio": 17
+ "preProcessors": [
+ {
+ "name": "GaussianBlur",
+ "options": {
+ "kSize": [
+ 3,
+ 3
+ ],
+ "sigmaX": 0
+ }
},
- "OverrideFlags": {
- "noCropping": true
+ {
+ "name": "CropPage",
+ "options": {
+ "morphKernel": [
+ 10,
+ 10
+ ]
+ }
}
- },
- "Concatenations": {
- "Roll": [
- "Squad",
- "Medium",
- "roll0",
- "roll1",
- "roll2",
- "roll3",
- "roll4",
- "roll5",
- "roll6",
- "roll7",
- "roll8"
- ],
- "q5": [
- "q5.1",
- "q5.2"
- ],
- "q6": [
- "q6.1",
- "q6.2"
- ],
- "q7": [
- "q7.1",
- "q7.2"
- ],
- "q9": [
- "q9.1",
- "q9.2"
- ],
- "q11": [
- "q8.1",
- "q8.2"
- ]
- },
- "Singles": [
- "q1",
- "q2",
- "q3",
- "q4",
- "q10",
- "q11",
- "q12",
- "q13",
- "q14",
- "q15",
- "q16",
- "q17",
- "q18",
- "q19",
- "q20"
],
- "QBlocks": {
- "Medium": {
- "qType": "QTYPE_MED",
- "orig": [
- 160,
- 285
- ],
- "bigGaps": [
- 115,
- 11
- ],
- "gaps": [
- 59,
- 46
- ],
- "qNos": [
- [
- [
- "Medium"
- ]
- ]
- ]
- },
- "Roll": {
- "qType": "QTYPE_ROLL",
- "orig": [
- 218,
- 285
- ],
- "bigGaps": [
- 115,
- 11
- ],
- "gaps": [
- 58,
- 46
- ],
- "qNos": [
- [
- [
- "roll0",
- "roll1",
- "roll2",
- "roll3",
- "roll4",
- "roll5",
- "roll6",
- "roll7",
- "roll8"
- ]
- ]
- ]
- },
- "Int1": {
- "qType": "QTYPE_INT",
- "orig": [
- 903,
- 285
- ],
- "bigGaps": [
- 128,
- 11
- ],
- "gaps": [
- 59,
- 46
- ],
- "qNos": [
- [
- [
- "q5.1",
- "q5.2"
- ],
- [
- "q6.1",
- "q6.2"
- ],
- [
- "q7.1",
- "q7.2"
- ]
- ]
- ]
- },
- "Int2": {
- "qType": "QTYPE_INT",
- "orig": [
- 1418,
- 285
- ],
- "bigGaps": [
- 128,
- 11
- ],
- "gaps": [
- 59,
- 46
- ],
- "qNos": [
- [
- [
- "q8.1",
- "q8.2"
- ],
- [
- "q9.1",
- "q9.2"
- ]
- ]
- ]
- },
- "Mcq1": {
- "qType": "QTYPE_MCQ4",
- "orig": [
- 118,
- 857
- ],
- "bigGaps": [
- 115,
- 181
- ],
- "gaps": [
- 59,
- 53
- ],
- "qNos": [
- [
- [
- "q1",
- "q2",
- "q3",
- "q4"
- ],
- [
- "q10",
- "q11",
- "q12",
- "q13"
- ]
- ]
- ]
- },
- "Mcq2": {
- "qType": "QTYPE_MCQ4",
- "orig": [
- 905,
- 860
- ],
- "bigGaps": [
- 115,
- 180
- ],
- "gaps": [
- 59,
- 53
- ],
- "qNos": [
- [
- [
- "q14",
- "q15",
- "q16"
- ]
- ]
- ]
- },
- "Mcq3": {
- "qType": "QTYPE_MCQ4",
- "orig": [
- 905,
- 1198
- ],
- "bigGaps": [
- 115,
- 180
- ],
- "gaps": [
- 59,
- 53
- ],
- "qNos": [
- [
- [
- "q17",
- "q18",
- "q19",
- "q20"
- ]
- ]
- ]
+ "fieldBlocks": {
+ "MCQBlock1": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 134,
+ 684
+ ],
+ "fieldLabels": [
+ "q1..11"
+ ],
+ "bubblesGap": 79,
+ "labelsGap": 62
}
}
-}
\ No newline at end of file
+}
diff --git a/samples/sample5/README.md b/samples/sample5/README.md
new file mode 100644
index 00000000..b5ed8ccb
--- /dev/null
+++ b/samples/sample5/README.md
@@ -0,0 +1,6 @@
+### OMRChecker Sample
+
+This sample demonstrates multiple things, namely -
+- Running OMRChecker on images scanned using popular document scanning apps
+- Using a common template.json file for sub-folders (e.g. multiple scan batches)
+- Using evaluation.json file with custom marking (without streak-based marking)
diff --git a/samples/sample3/CamScanner/sheet2.jpg b/samples/sample5/ScanBatch1/camscanner-1.jpg
similarity index 100%
rename from samples/sample3/CamScanner/sheet2.jpg
rename to samples/sample5/ScanBatch1/camscanner-1.jpg
diff --git a/samples/sample3/CamScanner/sheet1.jpg b/samples/sample5/ScanBatch2/camscanner-2.jpg
similarity index 100%
rename from samples/sample3/CamScanner/sheet1.jpg
rename to samples/sample5/ScanBatch2/camscanner-2.jpg
diff --git a/samples/sample5/config.json b/samples/sample5/config.json
new file mode 100644
index 00000000..a9daf449
--- /dev/null
+++ b/samples/sample5/config.json
@@ -0,0 +1,11 @@
+{
+ "dimensions": {
+ "display_height": 2480,
+ "display_width": 1640,
+ "processing_height": 820,
+ "processing_width": 666
+ },
+ "outputs": {
+ "show_image_level": 5
+ }
+}
diff --git a/samples/sample5/evaluation.json b/samples/sample5/evaluation.json
new file mode 100644
index 00000000..3332fe6a
--- /dev/null
+++ b/samples/sample5/evaluation.json
@@ -0,0 +1,93 @@
+{
+ "source_type": "custom",
+ "options": {
+ "questions_in_order": [
+ "q1..22"
+ ],
+ "answers_in_order": [
+ "C",
+ "C",
+ "B",
+ "C",
+ "C",
+ [
+ "1",
+ "01"
+ ],
+ "19",
+ "10",
+ "10",
+ "18",
+ "D",
+ "A",
+ "D",
+ "D",
+ "D",
+ "C",
+ "C",
+ "C",
+ "C",
+ "D",
+ "B",
+ "A"
+ ],
+ "should_explain_scoring": true
+ },
+ "marking_schemes": {
+ "DEFAULT": {
+ "correct": "1",
+ "incorrect": "0",
+ "unmarked": "0"
+ },
+ "BOOMERANG_1": {
+ "questions": [
+ "q1..5"
+ ],
+ "marking": {
+ "correct": 4,
+ "incorrect": -1,
+ "unmarked": 0
+ }
+ },
+ "PROXIMITY_1": {
+ "questions": [
+ "q6..10"
+ ],
+ "marking": {
+ "correct": 3,
+ "incorrect": -1,
+ "unmarked": 0
+ }
+ },
+ "FIBONACCI_SECTION_1": {
+ "questions": [
+ "q11..14"
+ ],
+ "marking": {
+ "correct": 2,
+ "incorrect": -1,
+ "unmarked": 0
+ }
+ },
+ "POWER_SECTION_1": {
+ "questions": [
+ "q15..18"
+ ],
+ "marking": {
+ "correct": 1,
+ "incorrect": 0,
+ "unmarked": 0
+ }
+ },
+ "FIBONACCI_SECTION_2": {
+ "questions": [
+ "q19..22"
+ ],
+ "marking": {
+ "correct": 2,
+ "incorrect": -1,
+ "unmarked": 0
+ }
+ }
+ }
+}
diff --git a/samples/sample5/omr_marker.jpg b/samples/sample5/omr_marker.jpg
new file mode 100644
index 00000000..0929feec
Binary files /dev/null and b/samples/sample5/omr_marker.jpg differ
diff --git a/samples/sample5/template.json b/samples/sample5/template.json
new file mode 100644
index 00000000..39f6e83d
--- /dev/null
+++ b/samples/sample5/template.json
@@ -0,0 +1,188 @@
+{
+ "pageDimensions": [
+ 1846,
+ 1500
+ ],
+ "bubbleDimensions": [
+ 40,
+ 40
+ ],
+ "preProcessors": [
+ {
+ "name": "CropOnMarkers",
+ "options": {
+ "relativePath": "omr_marker.jpg",
+ "sheetToMarkerWidthRatio": 17
+ }
+ }
+ ],
+ "customLabels": {
+ "Roll": [
+ "Medium",
+ "roll1..9"
+ ],
+ "q6": [
+ "q6_1",
+ "q6_2"
+ ],
+ "q7": [
+ "q7_1",
+ "q7_2"
+ ],
+ "q8": [
+ "q8_1",
+ "q8_2"
+ ],
+ "q9": [
+ "q9_1",
+ "q9_2"
+ ],
+ "q10": [
+ "q10_1",
+ "q10_2"
+ ]
+ },
+ "fieldBlocks": {
+ "Medium": {
+ "bubbleValues": [
+ "E",
+ "H"
+ ],
+ "direction": "vertical",
+ "origin": [
+ 200,
+ 215
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 0,
+ "fieldLabels": [
+ "Medium"
+ ]
+ },
+ "Roll": {
+ "fieldType": "QTYPE_INT",
+ "origin": [
+ 261,
+ 210
+ ],
+ "bubblesGap": 46.5,
+ "labelsGap": 58,
+ "fieldLabels": [
+ "roll1..9"
+ ]
+ },
+ "Int1": {
+ "fieldType": "QTYPE_INT",
+ "origin": [
+ 935,
+ 211
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 57,
+ "fieldLabels": [
+ "q6_1",
+ "q6_2"
+ ]
+ },
+ "Int2": {
+ "fieldType": "QTYPE_INT",
+ "origin": [
+ 1100,
+ 211
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 57,
+ "fieldLabels": [
+ "q7_1",
+ "q7_2"
+ ]
+ },
+ "Int3": {
+ "fieldType": "QTYPE_INT",
+ "origin": [
+ 1275,
+ 211
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 57,
+ "fieldLabels": [
+ "q8_1",
+ "q8_2"
+ ]
+ },
+ "Int4": {
+ "fieldType": "QTYPE_INT",
+ "origin": [
+ 1449,
+ 211
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 57,
+ "fieldLabels": [
+ "q9_1",
+ "q9_2"
+ ]
+ },
+ "Int5": {
+ "fieldType": "QTYPE_INT",
+ "origin": [
+ 1620,
+ 211
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 57,
+ "fieldLabels": [
+ "q10_1",
+ "q10_2"
+ ]
+ },
+ "Mcq1": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 198,
+ 826
+ ],
+ "bubblesGap": 93,
+ "labelsGap": 62,
+ "fieldLabels": [
+ "q1..5"
+ ]
+ },
+ "Mcq2": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 833,
+ 830
+ ],
+ "bubblesGap": 71,
+ "labelsGap": 61,
+ "fieldLabels": [
+ "q11..14"
+ ]
+ },
+ "Mcq3": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 833,
+ 1270
+ ],
+ "bubblesGap": 71,
+ "labelsGap": 61,
+ "fieldLabels": [
+ "q15..18"
+ ]
+ },
+ "Mcq4": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 1481,
+ 830
+ ],
+ "bubblesGap": 73,
+ "labelsGap": 61,
+ "fieldLabels": [
+ "q19..22"
+ ]
+ }
+ }
+}
diff --git a/samples/sample6/config.json b/samples/sample6/config.json
new file mode 100644
index 00000000..6f39f0ca
--- /dev/null
+++ b/samples/sample6/config.json
@@ -0,0 +1,11 @@
+{
+ "dimensions": {
+ "display_width": 2480,
+ "display_height": 3508,
+ "processing_width": 1653,
+ "processing_height": 2339
+ },
+ "outputs": {
+ "show_image_level": 5
+ }
+}
diff --git a/samples/sample6/doc-scans/sample_roll_01.jpg b/samples/sample6/doc-scans/sample_roll_01.jpg
new file mode 100644
index 00000000..25093a37
Binary files /dev/null and b/samples/sample6/doc-scans/sample_roll_01.jpg differ
diff --git a/samples/sample6/doc-scans/sample_roll_02.jpg b/samples/sample6/doc-scans/sample_roll_02.jpg
new file mode 100644
index 00000000..a81ee4fc
Binary files /dev/null and b/samples/sample6/doc-scans/sample_roll_02.jpg differ
diff --git a/samples/sample6/doc-scans/sample_roll_03.jpg b/samples/sample6/doc-scans/sample_roll_03.jpg
new file mode 100644
index 00000000..94296f4e
Binary files /dev/null and b/samples/sample6/doc-scans/sample_roll_03.jpg differ
diff --git a/samples/sample6/readme.md b/samples/sample6/readme.md
new file mode 100644
index 00000000..2a02ee90
--- /dev/null
+++ b/samples/sample6/readme.md
@@ -0,0 +1,21 @@
+# Demo for feature-based alignment
+
+## Background
+OMR is used to match student roll on final exam scripts. Scripts are scanned using a document scanner and the cover pages are extracted for OMR. Even though a document scanner does not produce any warpped perspective, the alignment is not perfect, causing some rotation and translation in the scans.
+
+The scripts in this sample were specifically selected incorrectly marked scripts to demonstrate how feature-based alignment can correct transformation errors using a reference image. In the actual batch. 156 out of 532 scripts were incorrectly marked. With feature-based alignment, all scripts were correctly marked.
+
+## Usage
+Two template files are given in the sample folder, one with feature-based alignment (template_fb_align), the other without (template_no_fb_align).
+
+## Additional Notes
+
+### Reference Image
+When using a reference image for feature-based alignment, it is better not to have many repeated patterns as it is causes ambiguity when trying to match similar feature points. The bubbles in an OMR form are identical and should not be used for feature-extraction.
+
+Thus, the reference image should be cleared of any bubbles. Forms with lots of text as in this example would be effective.
+
+Note the reference image in this sample was generated from a vector pdf, and not from a scanned blank, producing in a perfectly aligned reference.
+
+### Level adjustment
+The bubbles on the scripts were not shaded dark enough. Thus, a level adjustment was done to bring the black point to 70% to darken the light shading. White point was brought down to 80% to remove the light-grey background in the columns.
diff --git a/samples/sample6/reference.png b/samples/sample6/reference.png
new file mode 100644
index 00000000..5351267f
Binary files /dev/null and b/samples/sample6/reference.png differ
diff --git a/samples/sample6/template.json b/samples/sample6/template.json
new file mode 100644
index 00000000..57605a6c
--- /dev/null
+++ b/samples/sample6/template.json
@@ -0,0 +1,110 @@
+{
+ "pageDimensions": [
+ 2480,
+ 3508
+ ],
+ "bubbleDimensions": [
+ 42,
+ 42
+ ],
+ "preProcessors": [
+ {
+ "name": "Levels",
+ "options": {
+ "low": 0.7,
+ "high": 0.8
+ }
+ },
+ {
+ "name": "GaussianBlur",
+ "options": {
+ "kSize": [
+ 3,
+ 3
+ ],
+ "sigmaX": 0
+ }
+ }
+ ],
+ "customLabels": {
+ "Roll": [
+ "stu",
+ "roll1..7",
+ "check_1",
+ "check_2"
+ ]
+ },
+ "fieldBlocks": {
+ "Check1": {
+ "origin": [
+ 2033,
+ 1290
+ ],
+ "bubblesGap": 50,
+ "labelsGap": 50,
+ "fieldLabels": [
+ "check_1"
+ ],
+ "bubbleValues": [
+ "A",
+ "B",
+ "E",
+ "H",
+ "J",
+ "L",
+ "M"
+ ],
+ "direction": "vertical"
+ },
+ "Check2": {
+ "origin": [
+ 2083,
+ 1290
+ ],
+ "bubblesGap": 50,
+ "labelsGap": 50,
+ "fieldLabels": [
+ "check_2"
+ ],
+ "bubbleValues": [
+ "N",
+ "R",
+ "U",
+ "W",
+ "X",
+ "Y"
+ ],
+ "direction": "vertical"
+ },
+ "Stu": {
+ "origin": [
+ 1636,
+ 1290
+ ],
+ "bubblesGap": 50,
+ "labelsGap": 50,
+ "fieldLabels": [
+ "stu"
+ ],
+ "bubbleValues": [
+ "U",
+ "A",
+ "HT",
+ "GT"
+ ],
+ "direction": "vertical"
+ },
+ "Roll": {
+ "fieldType": "QTYPE_INT",
+ "origin": [
+ 1685,
+ 1290
+ ],
+ "bubblesGap": 50.5,
+ "labelsGap": 50.5,
+ "fieldLabels": [
+ "roll1..7"
+ ]
+ }
+ }
+}
diff --git a/samples/sample6/template_fb_align.json b/samples/sample6/template_fb_align.json
new file mode 100644
index 00000000..7c0c2397
--- /dev/null
+++ b/samples/sample6/template_fb_align.json
@@ -0,0 +1,118 @@
+{
+ "pageDimensions": [
+ 2480,
+ 3508
+ ],
+ "bubbleDimensions": [
+ 42,
+ 42
+ ],
+ "preProcessors": [
+ {
+ "name": "Levels",
+ "options": {
+ "low": 0.7,
+ "high": 0.8
+ }
+ },
+ {
+ "name": "FeatureBasedAlignment",
+ "options": {
+ "reference": "reference.png",
+ "maxFeatures": 1000,
+ "2d": true
+ }
+ },
+ {
+ "name": "GaussianBlur",
+ "options": {
+ "kSize": [
+ 3,
+ 3
+ ],
+ "sigmaX": 0
+ }
+ }
+ ],
+ "customLabels": {
+ "Roll": [
+ "stu",
+ "roll1..7",
+ "check_1",
+ "check_2"
+ ]
+ },
+ "fieldBlocks": {
+ "Check1": {
+ "origin": [
+ 2033,
+ 1290
+ ],
+ "bubblesGap": 50,
+ "labelsGap": 50,
+ "fieldLabels": [
+ "check_1"
+ ],
+ "bubbleValues": [
+ "A",
+ "B",
+ "E",
+ "H",
+ "J",
+ "L",
+ "M"
+ ],
+ "direction": "vertical"
+ },
+ "Check2": {
+ "origin": [
+ 2083,
+ 1290
+ ],
+ "bubblesGap": 50,
+ "labelsGap": 50,
+ "fieldLabels": [
+ "check_2"
+ ],
+ "bubbleValues": [
+ "N",
+ "R",
+ "U",
+ "W",
+ "X",
+ "Y"
+ ],
+ "direction": "vertical"
+ },
+ "Stu": {
+ "origin": [
+ 1636,
+ 1290
+ ],
+ "bubblesGap": 50,
+ "labelsGap": 50,
+ "fieldLabels": [
+ "stu"
+ ],
+ "bubbleValues": [
+ "U",
+ "A",
+ "HT",
+ "GT"
+ ],
+ "direction": "vertical"
+ },
+ "Roll": {
+ "fieldType": "QTYPE_INT",
+ "origin": [
+ 1685,
+ 1290
+ ],
+ "bubblesGap": 50.5,
+ "labelsGap": 50.5,
+ "fieldLabels": [
+ "roll1..7"
+ ]
+ }
+ }
+}
diff --git a/samples/sample6/template_no_fb_align.json b/samples/sample6/template_no_fb_align.json
new file mode 100644
index 00000000..57605a6c
--- /dev/null
+++ b/samples/sample6/template_no_fb_align.json
@@ -0,0 +1,110 @@
+{
+ "pageDimensions": [
+ 2480,
+ 3508
+ ],
+ "bubbleDimensions": [
+ 42,
+ 42
+ ],
+ "preProcessors": [
+ {
+ "name": "Levels",
+ "options": {
+ "low": 0.7,
+ "high": 0.8
+ }
+ },
+ {
+ "name": "GaussianBlur",
+ "options": {
+ "kSize": [
+ 3,
+ 3
+ ],
+ "sigmaX": 0
+ }
+ }
+ ],
+ "customLabels": {
+ "Roll": [
+ "stu",
+ "roll1..7",
+ "check_1",
+ "check_2"
+ ]
+ },
+ "fieldBlocks": {
+ "Check1": {
+ "origin": [
+ 2033,
+ 1290
+ ],
+ "bubblesGap": 50,
+ "labelsGap": 50,
+ "fieldLabels": [
+ "check_1"
+ ],
+ "bubbleValues": [
+ "A",
+ "B",
+ "E",
+ "H",
+ "J",
+ "L",
+ "M"
+ ],
+ "direction": "vertical"
+ },
+ "Check2": {
+ "origin": [
+ 2083,
+ 1290
+ ],
+ "bubblesGap": 50,
+ "labelsGap": 50,
+ "fieldLabels": [
+ "check_2"
+ ],
+ "bubbleValues": [
+ "N",
+ "R",
+ "U",
+ "W",
+ "X",
+ "Y"
+ ],
+ "direction": "vertical"
+ },
+ "Stu": {
+ "origin": [
+ 1636,
+ 1290
+ ],
+ "bubblesGap": 50,
+ "labelsGap": 50,
+ "fieldLabels": [
+ "stu"
+ ],
+ "bubbleValues": [
+ "U",
+ "A",
+ "HT",
+ "GT"
+ ],
+ "direction": "vertical"
+ },
+ "Roll": {
+ "fieldType": "QTYPE_INT",
+ "origin": [
+ 1685,
+ 1290
+ ],
+ "bubblesGap": 50.5,
+ "labelsGap": 50.5,
+ "fieldLabels": [
+ "roll1..7"
+ ]
+ }
+ }
+}
diff --git a/src/__init__.py b/src/__init__.py
new file mode 100644
index 00000000..7fc528b4
--- /dev/null
+++ b/src/__init__.py
@@ -0,0 +1,5 @@
+# https://docs.python.org/3/tutorial/modules.html#:~:text=The%20__init__.py,on%20the%20module%20search%20path.
+from src.logger import logger
+
+# It takes a few seconds for the imports
+logger.info(f"Loading OMRChecker modules...")
diff --git a/src/constants.py b/src/constants.py
new file mode 100644
index 00000000..f1cdc87b
--- /dev/null
+++ b/src/constants.py
@@ -0,0 +1,54 @@
+"""
+
+ OMRChecker
+
+ Author: Udayraj Deshmukh
+ Github: https://github.com/Udayraj123
+
+"""
+from dotmap import DotMap
+
+# Filenames
+TEMPLATE_FILENAME = "template.json"
+EVALUATION_FILENAME = "evaluation.json"
+CONFIG_FILENAME = "config.json"
+
+FIELD_LABEL_NUMBER_REGEX = r"([^\d]+)(\d*)"
+#
+ERROR_CODES = DotMap(
+ {
+ "MULTI_BUBBLE_WARN": 1,
+ "NO_MARKER_ERR": 2,
+ },
+ _dynamic=False,
+)
+
+FIELD_TYPES = {
+ "QTYPE_INT": {
+ "bubbleValues": ["0", "1", "2", "3", "4", "5", "6", "7", "8", "9"],
+ "direction": "vertical",
+ },
+ "QTYPE_INT_FROM_1": {
+ "bubbleValues": ["1", "2", "3", "4", "5", "6", "7", "8", "9", "0"],
+ "direction": "vertical",
+ },
+ "QTYPE_MCQ4": {"bubbleValues": ["A", "B", "C", "D"], "direction": "horizontal"},
+ "QTYPE_MCQ5": {
+ "bubbleValues": ["A", "B", "C", "D", "E"],
+ "direction": "horizontal",
+ },
+ #
+ # You can create and append custom field types here-
+ #
+}
+
+# TODO: move to interaction.py
+TEXT_SIZE = 0.95
+CLR_BLACK = (50, 150, 150)
+CLR_WHITE = (250, 250, 250)
+CLR_GRAY = (130, 130, 130)
+CLR_DARK_GRAY = (100, 100, 100)
+
+# TODO: move to config.json
+GLOBAL_PAGE_THRESHOLD_WHITE = 200
+GLOBAL_PAGE_THRESHOLD_BLACK = 100
diff --git a/src/core.py b/src/core.py
new file mode 100644
index 00000000..d475105f
--- /dev/null
+++ b/src/core.py
@@ -0,0 +1,721 @@
+import os
+from collections import defaultdict
+from typing import Any
+
+import cv2
+import matplotlib.pyplot as plt
+import numpy as np
+
+import src.constants as constants
+from src.logger import logger
+from src.utils.image import CLAHE_HELPER, ImageUtils
+from src.utils.interaction import InteractionUtils
+
+
+class ImageInstanceOps:
+ """Class to hold fine-tuned utilities for a group of images. One instance for each processing directory."""
+
+ save_img_list: Any = defaultdict(list)
+
+ def __init__(self, tuning_config):
+ super().__init__()
+ self.tuning_config = tuning_config
+ self.save_image_level = tuning_config.outputs.save_image_level
+
+ def apply_preprocessors(self, file_path, in_omr, template):
+ tuning_config = self.tuning_config
+ # resize to conform to template
+ in_omr = ImageUtils.resize_util(
+ in_omr,
+ tuning_config.dimensions.processing_width,
+ tuning_config.dimensions.processing_height,
+ )
+
+ # run pre_processors in sequence
+ for pre_processor in template.pre_processors:
+ in_omr = pre_processor.apply_filter(in_omr, file_path)
+ return in_omr
+
+ def read_omr_response(self, template, image, name, save_dir=None):
+ config = self.tuning_config
+ auto_align = config.alignment_params.auto_align
+ try:
+ img = image.copy()
+ # origDim = img.shape[:2]
+ img = ImageUtils.resize_util(
+ img, template.page_dimensions[0], template.page_dimensions[1]
+ )
+ if img.max() > img.min():
+ img = ImageUtils.normalize_util(img)
+ # Processing copies
+ transp_layer = img.copy()
+ final_marked = img.copy()
+
+ morph = img.copy()
+ self.append_save_img(3, morph)
+
+ if auto_align:
+ # Note: clahe is good for morphology, bad for thresholding
+ morph = CLAHE_HELPER.apply(morph)
+ self.append_save_img(3, morph)
+ # Remove shadows further, make columns/boxes darker (less gamma)
+ morph = ImageUtils.adjust_gamma(
+ morph, config.threshold_params.GAMMA_LOW
+ )
+ # TODO: all numbers should come from either constants or config
+ _, morph = cv2.threshold(morph, 220, 220, cv2.THRESH_TRUNC)
+ morph = ImageUtils.normalize_util(morph)
+ self.append_save_img(3, morph)
+ if config.outputs.show_image_level >= 4:
+ InteractionUtils.show("morph1", morph, 0, 1, config)
+
+ # Move them to data class if needed
+ # Overlay Transparencies
+ alpha = 0.65
+ omr_response = {}
+ multi_marked, multi_roll = 0, 0
+
+ # TODO Make this part useful for visualizing status checks
+ # blackVals=[0]
+ # whiteVals=[255]
+
+ if config.outputs.show_image_level >= 5:
+ all_c_box_vals = {"int": [], "mcq": []}
+ # TODO: simplify this logic
+ q_nums = {"int": [], "mcq": []}
+
+ # Find Shifts for the field_blocks --> Before calculating threshold!
+ if auto_align:
+ # print("Begin Alignment")
+ # Open : erode then dilate
+ v_kernel = cv2.getStructuringElement(cv2.MORPH_RECT, (2, 10))
+ morph_v = cv2.morphologyEx(
+ morph, cv2.MORPH_OPEN, v_kernel, iterations=3
+ )
+ _, morph_v = cv2.threshold(morph_v, 200, 200, cv2.THRESH_TRUNC)
+ morph_v = 255 - ImageUtils.normalize_util(morph_v)
+
+ if config.outputs.show_image_level >= 3:
+ InteractionUtils.show(
+ "morphed_vertical", morph_v, 0, 1, config=config
+ )
+
+ # InteractionUtils.show("morph1",morph,0,1,config=config)
+ # InteractionUtils.show("morphed_vertical",morph_v,0,1,config=config)
+
+ self.append_save_img(3, morph_v)
+
+ morph_thr = 60 # for Mobile images, 40 for scanned Images
+ _, morph_v = cv2.threshold(morph_v, morph_thr, 255, cv2.THRESH_BINARY)
+ # kernel best tuned to 5x5 now
+ morph_v = cv2.erode(morph_v, np.ones((5, 5), np.uint8), iterations=2)
+
+ self.append_save_img(3, morph_v)
+ # h_kernel = cv2.getStructuringElement(cv2.MORPH_RECT, (10, 2))
+ # morph_h = cv2.morphologyEx(morph, cv2.MORPH_OPEN, h_kernel, iterations=3)
+ # ret, morph_h = cv2.threshold(morph_h,200,200,cv2.THRESH_TRUNC)
+ # morph_h = 255 - normalize_util(morph_h)
+ # InteractionUtils.show("morph_h",morph_h,0,1,config=config)
+ # _, morph_h = cv2.threshold(morph_h,morph_thr,255,cv2.THRESH_BINARY)
+ # morph_h = cv2.erode(morph_h, np.ones((5,5),np.uint8), iterations = 2)
+ if config.outputs.show_image_level >= 3:
+ InteractionUtils.show(
+ "morph_thr_eroded", morph_v, 0, 1, config=config
+ )
+
+ self.append_save_img(6, morph_v)
+
+ # template relative alignment code
+ for field_block in template.field_blocks:
+ s, d = field_block.origin, field_block.dimensions
+
+ match_col, max_steps, align_stride, thk = map(
+ config.alignment_params.get,
+ [
+ "match_col",
+ "max_steps",
+ "stride",
+ "thickness",
+ ],
+ )
+ shift, steps = 0, 0
+ while steps < max_steps:
+ left_mean = np.mean(
+ morph_v[
+ s[1] : s[1] + d[1],
+ s[0] + shift - thk : -thk + s[0] + shift + match_col,
+ ]
+ )
+ right_mean = np.mean(
+ morph_v[
+ s[1] : s[1] + d[1],
+ s[0]
+ + shift
+ - match_col
+ + d[0]
+ + thk : thk
+ + s[0]
+ + shift
+ + d[0],
+ ]
+ )
+
+ # For demonstration purposes-
+ # if(field_block.name == "int1"):
+ # ret = morph_v.copy()
+ # cv2.rectangle(ret,
+ # (s[0]+shift-thk,s[1]),
+ # (s[0]+shift+thk+d[0],s[1]+d[1]),
+ # constants.CLR_WHITE,
+ # 3)
+ # appendSaveImg(6,ret)
+ # print(shift, left_mean, right_mean)
+ left_shift, right_shift = left_mean > 100, right_mean > 100
+ if left_shift:
+ if right_shift:
+ break
+ else:
+ shift -= align_stride
+ else:
+ if right_shift:
+ shift += align_stride
+ else:
+ break
+ steps += 1
+
+ field_block.shift = shift
+ # print("Aligned field_block: ",field_block.name,"Corrected Shift:",
+ # field_block.shift,", dimensions:", field_block.dimensions,
+ # "origin:", field_block.origin,'\n')
+ # print("End Alignment")
+
+ final_align = None
+ if config.outputs.show_image_level >= 2:
+ initial_align = self.draw_template_layout(img, template, shifted=False)
+ final_align = self.draw_template_layout(
+ img, template, shifted=True, draw_qvals=True
+ )
+ # appendSaveImg(4,mean_vals)
+ self.append_save_img(2, initial_align)
+ self.append_save_img(2, final_align)
+
+ if auto_align:
+ final_align = np.hstack((initial_align, final_align))
+ self.append_save_img(5, img)
+
+ # Get mean bubbleValues n other stats
+ all_q_vals, all_q_strip_arrs, all_q_std_vals = [], [], []
+ total_q_strip_no = 0
+ for field_block in template.field_blocks:
+ box_w, box_h = field_block.bubble_dimensions
+ q_std_vals = []
+ for field_block_bubbles in field_block.traverse_bubbles:
+ q_strip_vals = []
+ for pt in field_block_bubbles:
+ # shifted
+ x, y = (pt.x + field_block.shift, pt.y)
+ rect = [y, y + box_h, x, x + box_w]
+ q_strip_vals.append(
+ cv2.mean(img[rect[0] : rect[1], rect[2] : rect[3]])[0]
+ # detectCross(img, rect) ? 100 : 0
+ )
+ q_std_vals.append(round(np.std(q_strip_vals), 2))
+ all_q_strip_arrs.append(q_strip_vals)
+ # _, _, _ = get_global_threshold(q_strip_vals, "QStrip Plot",
+ # plot_show=False, sort_in_plot=True)
+ # hist = getPlotImg()
+ # InteractionUtils.show("QStrip "+field_block_bubbles[0].field_label, hist, 0, 1,config=config)
+ all_q_vals.extend(q_strip_vals)
+ # print(total_q_strip_no, field_block_bubbles[0].field_label, q_std_vals[len(q_std_vals)-1])
+ total_q_strip_no += 1
+ all_q_std_vals.extend(q_std_vals)
+
+ global_std_thresh, _, _ = self.get_global_threshold(
+ all_q_std_vals
+ ) # , "Q-wise Std-dev Plot", plot_show=True, sort_in_plot=True)
+ # plt.show()
+ # hist = getPlotImg()
+ # InteractionUtils.show("StdHist", hist, 0, 1,config=config)
+
+ # Note: Plotting takes Significant times here --> Change Plotting args
+ # to support show_image_level
+ # , "Mean Intensity Histogram",plot_show=True, sort_in_plot=True)
+ global_thr, _, _ = self.get_global_threshold(all_q_vals, looseness=4)
+
+ logger.info(
+ f"Thresholding: \tglobal_thr: {round(global_thr, 2)} \tglobal_std_THR: {round(global_std_thresh, 2)}\t{'(Looks like a Xeroxed OMR)' if (global_thr == 255) else ''}"
+ )
+ # plt.show()
+ # hist = getPlotImg()
+ # InteractionUtils.show("StdHist", hist, 0, 1,config=config)
+
+ # if(config.outputs.show_image_level>=1):
+ # hist = getPlotImg()
+ # InteractionUtils.show("Hist", hist, 0, 1,config=config)
+ # appendSaveImg(4,hist)
+ # appendSaveImg(5,hist)
+ # appendSaveImg(2,hist)
+
+ per_omr_threshold_avg, total_q_strip_no, total_q_box_no = 0, 0, 0
+ for field_block in template.field_blocks:
+ block_q_strip_no = 1
+ box_w, box_h = field_block.bubble_dimensions
+ shift = field_block.shift
+ s, d = field_block.origin, field_block.dimensions
+ key = field_block.name[:3]
+ # cv2.rectangle(final_marked,(s[0]+shift,s[1]),(s[0]+shift+d[0],
+ # s[1]+d[1]),CLR_BLACK,3)
+ for field_block_bubbles in field_block.traverse_bubbles:
+ # All Black or All White case
+ no_outliers = all_q_std_vals[total_q_strip_no] < global_std_thresh
+ # print(total_q_strip_no, field_block_bubbles[0].field_label,
+ # all_q_std_vals[total_q_strip_no], "no_outliers:", no_outliers)
+ per_q_strip_threshold = self.get_local_threshold(
+ all_q_strip_arrs[total_q_strip_no],
+ global_thr,
+ no_outliers,
+ f"Mean Intensity Histogram for {key}.{field_block_bubbles[0].field_label}.{block_q_strip_no}",
+ config.outputs.show_image_level >= 6,
+ )
+ # print(field_block_bubbles[0].field_label,key,block_q_strip_no, "THR: ",
+ # round(per_q_strip_threshold,2))
+ per_omr_threshold_avg += per_q_strip_threshold
+
+ # Note: Little debugging visualization - view the particular Qstrip
+ # if(
+ # 0
+ # # or "q17" in (field_block_bubbles[0].field_label)
+ # # or (field_block_bubbles[0].field_label+str(block_q_strip_no))=="q15"
+ # ):
+ # st, end = qStrip
+ # InteractionUtils.show("QStrip: "+key+"-"+str(block_q_strip_no),
+ # img[st[1] : end[1], st[0]+shift : end[0]+shift],0,config=config)
+
+ # TODO: get rid of total_q_box_no
+ detected_bubbles = []
+ for bubble in field_block_bubbles:
+ bubble_is_marked = (
+ per_q_strip_threshold > all_q_vals[total_q_box_no]
+ )
+ total_q_box_no += 1
+ if bubble_is_marked:
+ detected_bubbles.append(bubble)
+ x, y, field_value = (
+ bubble.x + field_block.shift,
+ bubble.y,
+ bubble.field_value,
+ )
+ cv2.rectangle(
+ final_marked,
+ (int(x + box_w / 12), int(y + box_h / 12)),
+ (
+ int(x + box_w - box_w / 12),
+ int(y + box_h - box_h / 12),
+ ),
+ constants.CLR_DARK_GRAY,
+ 3,
+ )
+
+ cv2.putText(
+ final_marked,
+ str(field_value),
+ (x, y),
+ cv2.FONT_HERSHEY_SIMPLEX,
+ constants.TEXT_SIZE,
+ (20, 20, 10),
+ int(1 + 3.5 * constants.TEXT_SIZE),
+ )
+ else:
+ cv2.rectangle(
+ final_marked,
+ (int(x + box_w / 10), int(y + box_h / 10)),
+ (
+ int(x + box_w - box_w / 10),
+ int(y + box_h - box_h / 10),
+ ),
+ constants.CLR_GRAY,
+ -1,
+ )
+
+ for bubble in detected_bubbles:
+ field_label, field_value = (
+ bubble.field_label,
+ bubble.field_value,
+ )
+ # Only send rolls multi-marked in the directory
+ multi_marked_local = field_label in omr_response
+ omr_response[field_label] = (
+ (omr_response[field_label] + field_value)
+ if multi_marked_local
+ else field_value
+ )
+ # TODO: generalize this into identifier
+ # multi_roll = multi_marked_local and "Roll" in str(q)
+ multi_marked = multi_marked or multi_marked_local
+
+ if len(detected_bubbles) == 0:
+ field_label = field_block_bubbles[0].field_label
+ omr_response[field_label] = field_block.empty_val
+
+ if config.outputs.show_image_level >= 5:
+ if key in all_c_box_vals:
+ q_nums[key].append(f"{key[:2]}_c{str(block_q_strip_no)}")
+ all_c_box_vals[key].append(
+ all_q_strip_arrs[total_q_strip_no]
+ )
+
+ block_q_strip_no += 1
+ total_q_strip_no += 1
+ # /for field_block
+
+ per_omr_threshold_avg /= total_q_strip_no
+ per_omr_threshold_avg = round(per_omr_threshold_avg, 2)
+ # Translucent
+ cv2.addWeighted(
+ final_marked, alpha, transp_layer, 1 - alpha, 0, final_marked
+ )
+ # Box types
+ if config.outputs.show_image_level >= 6:
+ # plt.draw()
+ f, axes = plt.subplots(len(all_c_box_vals), sharey=True)
+ f.canvas.manager.set_window_title(name)
+ ctr = 0
+ type_name = {
+ "int": "Integer",
+ "mcq": "MCQ",
+ "med": "MED",
+ "rol": "Roll",
+ }
+ for k, boxvals in all_c_box_vals.items():
+ axes[ctr].title.set_text(type_name[k] + " Type")
+ axes[ctr].boxplot(boxvals)
+ # thrline=axes[ctr].axhline(per_omr_threshold_avg,color='red',ls='--')
+ # thrline.set_label("Average THR")
+ axes[ctr].set_ylabel("Intensity")
+ axes[ctr].set_xticklabels(q_nums[k])
+ # axes[ctr].legend()
+ ctr += 1
+ # imshow will do the waiting
+ plt.tight_layout(pad=0.5)
+ plt.show()
+
+ if config.outputs.show_image_level >= 3 and final_align is not None:
+ final_align = ImageUtils.resize_util_h(
+ final_align, int(config.dimensions.display_height)
+ )
+ # [final_align.shape[1],0])
+ InteractionUtils.show(
+ "Template Alignment Adjustment", final_align, 0, 0, config=config
+ )
+
+ if config.outputs.save_detections and save_dir is not None:
+ if multi_roll:
+ save_dir = save_dir.joinpath("_MULTI_")
+ image_path = str(save_dir.joinpath(name))
+ ImageUtils.save_img(image_path, final_marked)
+
+ self.append_save_img(2, final_marked)
+
+ if save_dir is not None:
+ for i in range(config.outputs.save_image_level):
+ self.save_image_stacks(i + 1, name, save_dir)
+
+ return omr_response, final_marked, multi_marked, multi_roll
+
+ except Exception as e:
+ raise e
+
+ @staticmethod
+ def draw_template_layout(img, template, shifted=True, draw_qvals=False, border=-1):
+ img = ImageUtils.resize_util(
+ img, template.page_dimensions[0], template.page_dimensions[1]
+ )
+ final_align = img.copy()
+ for field_block in template.field_blocks:
+ s, d = field_block.origin, field_block.dimensions
+ box_w, box_h = field_block.bubble_dimensions
+ shift = field_block.shift
+ if shifted:
+ cv2.rectangle(
+ final_align,
+ (s[0] + shift, s[1]),
+ (s[0] + shift + d[0], s[1] + d[1]),
+ constants.CLR_BLACK,
+ 3,
+ )
+ else:
+ cv2.rectangle(
+ final_align,
+ (s[0], s[1]),
+ (s[0] + d[0], s[1] + d[1]),
+ constants.CLR_BLACK,
+ 3,
+ )
+ for field_block_bubbles in field_block.traverse_bubbles:
+ for pt in field_block_bubbles:
+ x, y = (pt.x + field_block.shift, pt.y) if shifted else (pt.x, pt.y)
+ cv2.rectangle(
+ final_align,
+ (int(x + box_w / 10), int(y + box_h / 10)),
+ (int(x + box_w - box_w / 10), int(y + box_h - box_h / 10)),
+ constants.CLR_GRAY,
+ border,
+ )
+ if draw_qvals:
+ rect = [y, y + box_h, x, x + box_w]
+ cv2.putText(
+ final_align,
+ f"{int(cv2.mean(img[rect[0] : rect[1], rect[2] : rect[3]])[0])}",
+ (rect[2] + 2, rect[0] + (box_h * 2) // 3),
+ cv2.FONT_HERSHEY_SIMPLEX,
+ 0.6,
+ constants.CLR_BLACK,
+ 2,
+ )
+ if shifted:
+ text_in_px = cv2.getTextSize(
+ field_block.name, cv2.FONT_HERSHEY_SIMPLEX, constants.TEXT_SIZE, 4
+ )
+ cv2.putText(
+ final_align,
+ field_block.name,
+ (int(s[0] + d[0] - text_in_px[0][0]), int(s[1] - text_in_px[0][1])),
+ cv2.FONT_HERSHEY_SIMPLEX,
+ constants.TEXT_SIZE,
+ constants.CLR_BLACK,
+ 4,
+ )
+ return final_align
+
+ def get_global_threshold(
+ self,
+ q_vals_orig,
+ plot_title=None,
+ plot_show=True,
+ sort_in_plot=True,
+ looseness=1,
+ ):
+ """
+ Note: Cannot assume qStrip has only-gray or only-white bg
+ (in which case there is only one jump).
+ So there will be either 1 or 2 jumps.
+ 1 Jump :
+ ......
+ ||||||
+ |||||| <-- risky THR
+ |||||| <-- safe THR
+ ....||||||
+ ||||||||||
+
+ 2 Jumps :
+ ......
+ |||||| <-- wrong THR
+ ....||||||
+ |||||||||| <-- safe THR
+ ..||||||||||
+ ||||||||||||
+
+ The abstract "First LARGE GAP" is perfect for this.
+ Current code is considering ONLY TOP 2 jumps(>= MIN_GAP) to be big,
+ gives the smaller one
+
+ """
+ config = self.tuning_config
+ PAGE_TYPE_FOR_THRESHOLD, MIN_JUMP, JUMP_DELTA = map(
+ config.threshold_params.get,
+ [
+ "PAGE_TYPE_FOR_THRESHOLD",
+ "MIN_JUMP",
+ "JUMP_DELTA",
+ ],
+ )
+
+ global_default_threshold = (
+ constants.GLOBAL_PAGE_THRESHOLD_WHITE
+ if PAGE_TYPE_FOR_THRESHOLD == "white"
+ else constants.GLOBAL_PAGE_THRESHOLD_BLACK
+ )
+
+ # Sort the Q bubbleValues
+ # TODO: Change var name of q_vals
+ q_vals = sorted(q_vals_orig)
+ # Find the FIRST LARGE GAP and set it as threshold:
+ ls = (looseness + 1) // 2
+ l = len(q_vals) - ls
+ max1, thr1 = MIN_JUMP, global_default_threshold
+ for i in range(ls, l):
+ jump = q_vals[i + ls] - q_vals[i - ls]
+ if jump > max1:
+ max1 = jump
+ thr1 = q_vals[i - ls] + jump / 2
+
+ # NOTE: thr2 is deprecated, thus is JUMP_DELTA
+ # Make use of the fact that the JUMP_DELTA(Vertical gap ofc) between
+ # values at detected jumps would be atleast 20
+ max2, thr2 = MIN_JUMP, global_default_threshold
+ # Requires atleast 1 gray box to be present (Roll field will ensure this)
+ for i in range(ls, l):
+ jump = q_vals[i + ls] - q_vals[i - ls]
+ new_thr = q_vals[i - ls] + jump / 2
+ if jump > max2 and abs(thr1 - new_thr) > JUMP_DELTA:
+ max2 = jump
+ thr2 = new_thr
+ # global_thr = min(thr1,thr2)
+ global_thr, j_low, j_high = thr1, thr1 - max1 // 2, thr1 + max1 // 2
+
+ # # For normal images
+ # thresholdRead = 116
+ # if(thr1 > thr2 and thr2 > thresholdRead):
+ # print("Note: taking safer thr line.")
+ # global_thr, j_low, j_high = thr2, thr2 - max2//2, thr2 + max2//2
+
+ if plot_title:
+ _, ax = plt.subplots()
+ ax.bar(range(len(q_vals_orig)), q_vals if sort_in_plot else q_vals_orig)
+ ax.set_title(plot_title)
+ thrline = ax.axhline(global_thr, color="green", ls="--", linewidth=5)
+ thrline.set_label("Global Threshold")
+ thrline = ax.axhline(thr2, color="red", ls=":", linewidth=3)
+ thrline.set_label("THR2 Line")
+ # thrline=ax.axhline(j_low,color='red',ls='-.', linewidth=3)
+ # thrline=ax.axhline(j_high,color='red',ls='-.', linewidth=3)
+ # thrline.set_label("Boundary Line")
+ # ax.set_ylabel("Mean Intensity")
+ ax.set_ylabel("Values")
+ ax.set_xlabel("Position")
+ ax.legend()
+ if plot_show:
+ plt.title(plot_title)
+ plt.show()
+
+ return global_thr, j_low, j_high
+
+ def get_local_threshold(
+ self, q_vals, global_thr, no_outliers, plot_title=None, plot_show=True
+ ):
+ """
+ TODO: Update this documentation too-
+ //No more - Assumption : Colwise background color is uniformly gray or white,
+ but not alternating. In this case there is atmost one jump.
+
+ 0 Jump :
+ <-- safe THR?
+ .......
+ ...|||||||
+ |||||||||| <-- safe THR?
+ // How to decide given range is above or below gray?
+ -> global q_vals shall absolutely help here. Just run same function
+ on total q_vals instead of colwise _//
+ How to decide it is this case of 0 jumps
+
+ 1 Jump :
+ ......
+ ||||||
+ |||||| <-- risky THR
+ |||||| <-- safe THR
+ ....||||||
+ ||||||||||
+
+ """
+ config = self.tuning_config
+ # Sort the Q bubbleValues
+ q_vals = sorted(q_vals)
+
+ # Small no of pts cases:
+ # base case: 1 or 2 pts
+ if len(q_vals) < 3:
+ thr1 = (
+ global_thr
+ if np.max(q_vals) - np.min(q_vals) < config.threshold_params.MIN_GAP
+ else np.mean(q_vals)
+ )
+ else:
+ # qmin, qmax, qmean, qstd = round(np.min(q_vals),2), round(np.max(q_vals),2),
+ # round(np.mean(q_vals),2), round(np.std(q_vals),2)
+ # GVals = [round(abs(q-qmean),2) for q in q_vals]
+ # gmean, gstd = round(np.mean(GVals),2), round(np.std(GVals),2)
+ # # DISCRETION: Pretty critical factor in reading response
+ # # Doesn't work well for small number of values.
+ # DISCRETION = 2.7 # 2.59 was closest hit, 3.0 is too far
+ # L2MaxGap = round(max([abs(g-gmean) for g in GVals]),2)
+ # if(L2MaxGap > DISCRETION*gstd):
+ # no_outliers = False
+
+ # # ^Stackoverflow method
+ # print(field_label, no_outliers,"qstd",round(np.std(q_vals),2), "gstd", gstd,
+ # "Gaps in gvals",sorted([round(abs(g-gmean),2) for g in GVals],reverse=True),
+ # '\t',round(DISCRETION*gstd,2), L2MaxGap)
+
+ # else:
+ # Find the LARGEST GAP and set it as threshold: //(FIRST LARGE GAP)
+ l = len(q_vals) - 1
+ max1, thr1 = config.threshold_params.MIN_JUMP, 255
+ for i in range(1, l):
+ jump = q_vals[i + 1] - q_vals[i - 1]
+ if jump > max1:
+ max1 = jump
+ thr1 = q_vals[i - 1] + jump / 2
+ # print(field_label,q_vals,max1)
+
+ confident_jump = (
+ config.threshold_params.MIN_JUMP
+ + config.threshold_params.CONFIDENT_SURPLUS
+ )
+ # If not confident, then only take help of global_thr
+ if max1 < confident_jump:
+ if no_outliers:
+ # All Black or All White case
+ thr1 = global_thr
+ else:
+ # TODO: Low confidence parameters here
+ pass
+
+ # if(thr1 == 255):
+ # print("Warning: threshold is unexpectedly 255! (Outlier Delta issue?)",plot_title)
+
+ # Make a common plot function to show local and global thresholds
+ if plot_show and plot_title is not None:
+ _, ax = plt.subplots()
+ ax.bar(range(len(q_vals)), q_vals)
+ thrline = ax.axhline(thr1, color="green", ls=("-."), linewidth=3)
+ thrline.set_label("Local Threshold")
+ thrline = ax.axhline(global_thr, color="red", ls=":", linewidth=5)
+ thrline.set_label("Global Threshold")
+ ax.set_title(plot_title)
+ ax.set_ylabel("Bubble Mean Intensity")
+ ax.set_xlabel("Bubble Number(sorted)")
+ ax.legend()
+ # TODO append QStrip to this plot-
+ # appendSaveImg(6,getPlotImg())
+ if plot_show:
+ plt.show()
+ return thr1
+
+ def append_save_img(self, key, img):
+ if self.save_image_level >= int(key):
+ self.save_img_list[key].append(img.copy())
+
+ def save_image_stacks(self, key, filename, save_dir):
+ config = self.tuning_config
+ if self.save_image_level >= int(key) and self.save_img_list[key] != []:
+ name = os.path.splitext(filename)[0]
+ result = np.hstack(
+ tuple(
+ [
+ ImageUtils.resize_util_h(img, config.dimensions.display_height)
+ for img in self.save_img_list[key]
+ ]
+ )
+ )
+ result = ImageUtils.resize_util(
+ result,
+ min(
+ len(self.save_img_list[key]) * config.dimensions.display_width // 3,
+ int(config.dimensions.display_width * 2.5),
+ ),
+ )
+ ImageUtils.save_img(f"{save_dir}stack/{name}_{str(key)}_stack.jpg", result)
+
+ def reset_all_save_img(self):
+ for i in range(self.save_image_level):
+ self.save_img_list[i + 1] = []
diff --git a/src/defaults/__init__.py b/src/defaults/__init__.py
new file mode 100644
index 00000000..62ed2e6f
--- /dev/null
+++ b/src/defaults/__init__.py
@@ -0,0 +1,5 @@
+# https://docs.python.org/3/tutorial/modules.html#:~:text=The%20__init__.py,on%20the%20module%20search%20path.
+# Use all imports relative to root directory
+# (https://chrisyeh96.github.io/2017/08/08/definitive-guide-python-imports.html)
+from src.defaults.config import * # NOQA
+from src.defaults.template import * # NOQA
diff --git a/src/defaults/config.py b/src/defaults/config.py
new file mode 100644
index 00000000..1c6cad3f
--- /dev/null
+++ b/src/defaults/config.py
@@ -0,0 +1,35 @@
+from dotmap import DotMap
+
+CONFIG_DEFAULTS = DotMap(
+ {
+ "dimensions": {
+ "display_height": 2480,
+ "display_width": 1640,
+ "processing_height": 820,
+ "processing_width": 666,
+ },
+ "threshold_params": {
+ "GAMMA_LOW": 0.7,
+ "MIN_GAP": 30,
+ "MIN_JUMP": 25,
+ "CONFIDENT_SURPLUS": 5,
+ "JUMP_DELTA": 30,
+ "PAGE_TYPE_FOR_THRESHOLD": "white",
+ },
+ "alignment_params": {
+ # Note: 'auto_align' enables automatic template alignment, use if the scans show slight misalignments.
+ "auto_align": False,
+ "match_col": 5,
+ "max_steps": 20,
+ "stride": 1,
+ "thickness": 3,
+ },
+ "outputs": {
+ "show_image_level": 0,
+ "save_image_level": 0,
+ "save_detections": True,
+ "filter_out_multimarked_files": False,
+ },
+ },
+ _dynamic=False,
+)
diff --git a/src/defaults/template.py b/src/defaults/template.py
new file mode 100644
index 00000000..d0a2a831
--- /dev/null
+++ b/src/defaults/template.py
@@ -0,0 +1,6 @@
+TEMPLATE_DEFAULTS = {
+ "preProcessors": [],
+ "emptyValue": "",
+ "customLabels": {},
+ "outputColumns": [],
+}
diff --git a/src/entry.py b/src/entry.py
new file mode 100644
index 00000000..5cfcfd95
--- /dev/null
+++ b/src/entry.py
@@ -0,0 +1,370 @@
+"""
+
+ OMRChecker
+
+ Author: Udayraj Deshmukh
+ Github: https://github.com/Udayraj123
+
+"""
+import os
+from csv import QUOTE_NONNUMERIC
+from pathlib import Path
+from time import time
+
+import cv2
+import pandas as pd
+from rich.table import Table
+
+from src import constants
+from src.defaults import CONFIG_DEFAULTS
+from src.evaluation import EvaluationConfig, evaluate_concatenated_response
+from src.logger import console, logger
+from src.template import Template
+from src.utils.file import Paths, setup_dirs_for_paths, setup_outputs_for_template
+from src.utils.image import ImageUtils
+from src.utils.interaction import InteractionUtils, Stats
+from src.utils.parsing import get_concatenated_response, open_config_with_defaults
+
+# Load processors
+STATS = Stats()
+
+
+def entry_point(input_dir, args):
+ if not os.path.exists(input_dir):
+ raise Exception(f"Given input directory does not exist: '{input_dir}'")
+ curr_dir = input_dir
+ return process_dir(input_dir, curr_dir, args)
+
+
+def print_config_summary(
+ curr_dir,
+ omr_files,
+ template,
+ tuning_config,
+ local_config_path,
+ evaluation_config,
+ args,
+):
+ logger.info("")
+ table = Table(title="Current Configurations", show_header=False, show_lines=False)
+ table.add_column("Key", style="cyan", no_wrap=True)
+ table.add_column("Value", style="magenta")
+ table.add_row("Directory Path", f"{curr_dir}")
+ table.add_row("Count of Images", f"{len(omr_files)}")
+ table.add_row("Set Layout Mode ", "ON" if args["setLayout"] else "OFF")
+ table.add_row(
+ "Markers Detection",
+ "ON" if "CropOnMarkers" in template.pre_processors else "OFF",
+ )
+ table.add_row("Auto Alignment", f"{tuning_config.alignment_params.auto_align}")
+ table.add_row("Detected Template Path", f"{template}")
+ if local_config_path:
+ table.add_row("Detected Local Config", f"{local_config_path}")
+ if evaluation_config:
+ table.add_row("Detected Evaluation Config", f"{evaluation_config}")
+
+ table.add_row(
+ "Detected pre-processors",
+ f"{[pp.__class__.__name__ for pp in template.pre_processors]}",
+ )
+ console.print(table, justify="center")
+
+
+def process_dir(
+ root_dir,
+ curr_dir,
+ args,
+ template=None,
+ tuning_config=CONFIG_DEFAULTS,
+ evaluation_config=None,
+):
+ # Update local tuning_config (in current recursion stack)
+ local_config_path = curr_dir.joinpath(constants.CONFIG_FILENAME)
+ if os.path.exists(local_config_path):
+ tuning_config = open_config_with_defaults(local_config_path)
+
+ # Update local template (in current recursion stack)
+ local_template_path = curr_dir.joinpath(constants.TEMPLATE_FILENAME)
+ local_template_exists = os.path.exists(local_template_path)
+ if local_template_exists:
+ template = Template(
+ local_template_path,
+ tuning_config,
+ )
+ # Look for subdirectories for processing
+ subdirs = [d for d in curr_dir.iterdir() if d.is_dir()]
+
+ output_dir = Path(args["output_dir"], curr_dir.relative_to(root_dir))
+ paths = Paths(output_dir)
+
+ # look for images in current dir to process
+ exts = ("*.[pP][nN][gG]", "*.[jJ][pP][gG]", "*.[jJ][pP][eE][gG]")
+ omr_files = sorted([f for ext in exts for f in curr_dir.glob(ext)])
+
+ # Exclude images (take union over all pre_processors)
+ excluded_files = []
+ if template:
+ for pp in template.pre_processors:
+ excluded_files.extend(Path(p) for p in pp.exclude_files())
+
+ local_evaluation_path = curr_dir.joinpath(constants.EVALUATION_FILENAME)
+ if not args["setLayout"] and os.path.exists(local_evaluation_path):
+ if not local_template_exists:
+ logger.warning(
+ f"Found an evaluation file without a parent template file: {local_evaluation_path}"
+ )
+ evaluation_config = EvaluationConfig(
+ curr_dir,
+ local_evaluation_path,
+ template,
+ tuning_config,
+ )
+
+ excluded_files.extend(
+ Path(exclude_file) for exclude_file in evaluation_config.get_exclude_files()
+ )
+
+ omr_files = [f for f in omr_files if f not in excluded_files]
+
+ if omr_files:
+ if not template:
+ logger.error(
+ f"Found images, but no template in the directory tree \
+ of '{curr_dir}'. \nPlace {constants.TEMPLATE_FILENAME} in the \
+ appropriate directory."
+ )
+ raise Exception(
+ f"No template file found in the directory tree of {curr_dir}"
+ )
+
+ setup_dirs_for_paths(paths)
+ outputs_namespace = setup_outputs_for_template(paths, template)
+
+ print_config_summary(
+ curr_dir,
+ omr_files,
+ template,
+ tuning_config,
+ local_config_path,
+ evaluation_config,
+ args,
+ )
+ if args["setLayout"]:
+ show_template_layouts(omr_files, template, tuning_config)
+ else:
+ process_files(
+ omr_files,
+ template,
+ tuning_config,
+ evaluation_config,
+ outputs_namespace,
+ )
+
+ elif not subdirs:
+ # Each subdirectory should have images or should be non-leaf
+ logger.info(
+ f"No valid images or sub-folders found in {curr_dir}.\
+ Empty directories not allowed."
+ )
+
+ # recursively process sub-folders
+ for d in subdirs:
+ process_dir(
+ root_dir,
+ d,
+ args,
+ template,
+ tuning_config,
+ evaluation_config,
+ )
+
+
+def show_template_layouts(omr_files, template, tuning_config):
+ for file_path in omr_files:
+ file_name = file_path.name
+ file_path = str(file_path)
+ in_omr = cv2.imread(file_path, cv2.IMREAD_GRAYSCALE)
+ in_omr = template.image_instance_ops.apply_preprocessors(
+ file_path, in_omr, template
+ )
+ template_layout = template.image_instance_ops.draw_template_layout(
+ in_omr, template, shifted=False, border=2
+ )
+ InteractionUtils.show(
+ f"Template Layout: {file_name}", template_layout, 1, 1, config=tuning_config
+ )
+
+
+def process_files(
+ omr_files,
+ template,
+ tuning_config,
+ evaluation_config,
+ outputs_namespace,
+):
+ start_time = int(time())
+ files_counter = 0
+ STATS.files_not_moved = 0
+
+ for file_path in omr_files:
+ files_counter += 1
+ file_name = file_path.name
+
+ in_omr = cv2.imread(str(file_path), cv2.IMREAD_GRAYSCALE)
+
+ logger.info("")
+ logger.info(
+ f"({files_counter}) Opening image: \t'{file_path}'\tResolution: {in_omr.shape}"
+ )
+
+ template.image_instance_ops.reset_all_save_img()
+
+ template.image_instance_ops.append_save_img(1, in_omr)
+
+ in_omr = template.image_instance_ops.apply_preprocessors(
+ file_path, in_omr, template
+ )
+
+ if in_omr is None:
+ # Error OMR case
+ new_file_path = outputs_namespace.paths.errors_dir.joinpath(file_name)
+ outputs_namespace.OUTPUT_SET.append(
+ [file_name] + outputs_namespace.empty_resp
+ )
+ if check_and_move(
+ constants.ERROR_CODES.NO_MARKER_ERR, file_path, new_file_path
+ ):
+ err_line = [
+ file_name,
+ file_path,
+ new_file_path,
+ "NA",
+ ] + outputs_namespace.empty_resp
+ pd.DataFrame(err_line, dtype=str).T.to_csv(
+ outputs_namespace.files_obj["Errors"],
+ mode="a",
+ quoting=QUOTE_NONNUMERIC,
+ header=False,
+ index=False,
+ )
+ continue
+
+ # uniquify
+ file_id = str(file_name)
+ save_dir = outputs_namespace.paths.save_marked_dir
+ (
+ response_dict,
+ final_marked,
+ multi_marked,
+ _,
+ ) = template.image_instance_ops.read_omr_response(
+ template, image=in_omr, name=file_id, save_dir=save_dir
+ )
+
+ # TODO: move inner try catch here
+ # concatenate roll nos, set unmarked responses, etc
+ omr_response = get_concatenated_response(response_dict, template)
+
+ if (
+ evaluation_config is None
+ or not evaluation_config.get_should_explain_scoring()
+ ):
+ logger.info(f"Read Response: \n{omr_response}")
+
+ score = 0
+ if evaluation_config is not None:
+ score = evaluate_concatenated_response(
+ omr_response, evaluation_config, file_path, outputs_namespace.paths.evaluation_dir
+ )
+ logger.info(
+ f"(/{files_counter}) Graded with score: {round(score, 2)}\t for file: '{file_id}'"
+ )
+ else:
+ logger.info(f"(/{files_counter}) Processed file: '{file_id}'")
+
+ if tuning_config.outputs.show_image_level >= 2:
+ InteractionUtils.show(
+ f"Final Marked Bubbles : '{file_id}'",
+ ImageUtils.resize_util_h(
+ final_marked, int(tuning_config.dimensions.display_height * 1.3)
+ ),
+ 1,
+ 1,
+ config=tuning_config,
+ )
+
+ resp_array = []
+ for k in template.output_columns:
+ resp_array.append(omr_response[k])
+
+ outputs_namespace.OUTPUT_SET.append([file_name] + resp_array)
+
+ if multi_marked == 0 or not tuning_config.outputs.filter_out_multimarked_files:
+ STATS.files_not_moved += 1
+ new_file_path = save_dir.joinpath(file_id)
+ # Enter into Results sheet-
+ results_line = [file_name, file_path, new_file_path, score] + resp_array
+ # Write/Append to results_line file(opened in append mode)
+ pd.DataFrame(results_line, dtype=str).T.to_csv(
+ outputs_namespace.files_obj["Results"],
+ mode="a",
+ quoting=QUOTE_NONNUMERIC,
+ header=False,
+ index=False,
+ )
+ else:
+ # multi_marked file
+ logger.info(f"[{files_counter}] Found multi-marked file: '{file_id}'")
+ new_file_path = outputs_namespace.paths.multi_marked_dir.joinpath(file_name)
+ if check_and_move(
+ constants.ERROR_CODES.MULTI_BUBBLE_WARN, file_path, new_file_path
+ ):
+ mm_line = [file_name, file_path, new_file_path, "NA"] + resp_array
+ pd.DataFrame(mm_line, dtype=str).T.to_csv(
+ outputs_namespace.files_obj["MultiMarked"],
+ mode="a",
+ quoting=QUOTE_NONNUMERIC,
+ header=False,
+ index=False,
+ )
+ # else:
+ # TODO: Add appropriate record handling here
+ # pass
+
+ print_stats(start_time, files_counter, tuning_config)
+
+
+def check_and_move(error_code, file_path, filepath2):
+ # TODO: fix file movement into error/multimarked/invalid etc again
+ STATS.files_not_moved += 1
+ return True
+
+
+def print_stats(start_time, files_counter, tuning_config):
+ time_checking = max(1, round(time() - start_time, 2))
+ log = logger.info
+ log("")
+ log(f"{'Total file(s) moved': <27}: {STATS.files_moved}")
+ log(f"{'Total file(s) not moved': <27}: {STATS.files_not_moved}")
+ log("--------------------------------")
+ log(
+ f"{'Total file(s) processed': <27}: {files_counter} ({'Sum Tallied!' if files_counter == (STATS.files_moved + STATS.files_not_moved) else 'Not Tallying!'})"
+ )
+
+ if tuning_config.outputs.show_image_level <= 0:
+ log(
+ f"\nFinished Checking {files_counter} file(s) in {round(time_checking, 1)} seconds i.e. ~{round(time_checking / 60, 1)} minute(s)."
+ )
+ log(
+ f"{'OMR Processing Rate': <27}: \t ~ {round(time_checking / files_counter, 2)} seconds/OMR"
+ )
+ log(
+ f"{'OMR Processing Speed': <27}: \t ~ {round((files_counter * 60) / time_checking, 2)} OMRs/minute"
+ )
+ else:
+ log(f"\n{'Total script time': <27}: {time_checking} seconds")
+
+ if tuning_config.outputs.show_image_level <= 1:
+ log(
+ "\nTip: To see some awesome visuals, open config.json and increase 'show_image_level'"
+ )
diff --git a/src/evaluation.py b/src/evaluation.py
new file mode 100644
index 00000000..0b567169
--- /dev/null
+++ b/src/evaluation.py
@@ -0,0 +1,546 @@
+import ast
+import os
+import re
+from copy import deepcopy
+from csv import QUOTE_NONNUMERIC
+
+import cv2
+import pandas as pd
+from rich.table import Table
+
+from src.logger import console, logger
+from src.schemas.constants import (
+ BONUS_SECTION_PREFIX,
+ DEFAULT_SECTION_KEY,
+ MARKING_VERDICT_TYPES,
+)
+from src.utils.parsing import (
+ get_concatenated_response,
+ open_evaluation_with_validation,
+ parse_fields,
+ parse_float_or_fraction,
+)
+
+
+class AnswerMatcher:
+ def __init__(self, answer_item, section_marking_scheme):
+ self.section_marking_scheme = section_marking_scheme
+ self.answer_item = answer_item
+ self.answer_type = self.validate_and_get_answer_type(answer_item)
+ self.set_defaults_from_scheme(section_marking_scheme)
+
+ @staticmethod
+ def is_a_marking_score(answer_element):
+ # Note: strict type checking is already done at schema validation level,
+ # Here we focus on overall struct type
+ return type(answer_element) == str or type(answer_element) == int
+
+ @staticmethod
+ def is_standard_answer(answer_element):
+ return type(answer_element) == str and len(answer_element) >= 1
+
+ def validate_and_get_answer_type(self, answer_item):
+ if self.is_standard_answer(answer_item):
+ return "standard"
+ elif type(answer_item) == list:
+ if (
+ # Array of answer elements: ['A', 'B', 'AB']
+ len(answer_item) >= 2
+ and all(
+ self.is_standard_answer(answers_or_score)
+ for answers_or_score in answer_item
+ )
+ ):
+ return "multiple-correct"
+ elif (
+ # Array of two-tuples: [['A', 1], ['B', 1], ['C', 3], ['AB', 2]]
+ len(answer_item) >= 1
+ and all(
+ type(answer_and_score) == list and len(answer_and_score) == 2
+ for answer_and_score in answer_item
+ )
+ and all(
+ self.is_standard_answer(allowed_answer)
+ and self.is_a_marking_score(answer_score)
+ for allowed_answer, answer_score in answer_item
+ )
+ ):
+ return "multiple-correct-weighted"
+
+ logger.critical(
+ f"Unable to determine answer type for answer item: {answer_item}"
+ )
+ raise Exception("Unable to determine answer type")
+
+ def set_defaults_from_scheme(self, section_marking_scheme):
+ answer_type = self.answer_type
+ self.empty_val = section_marking_scheme.empty_val
+ answer_item = self.answer_item
+ self.marking = deepcopy(section_marking_scheme.marking)
+ # TODO: reuse part of parse_scheme_marking here -
+ if answer_type == "standard":
+ # no local overrides
+ pass
+ elif answer_type == "multiple-correct":
+ # override marking scheme scores for each allowed answer
+ for allowed_answer in answer_item:
+ self.marking[f"correct-{allowed_answer}"] = self.marking["correct"]
+ elif answer_type == "multiple-correct-weighted":
+ # Note: No override using marking scheme as answer scores are provided in answer_item
+ for allowed_answer, answer_score in answer_item:
+ self.marking[f"correct-{allowed_answer}"] = parse_float_or_fraction(
+ answer_score
+ )
+
+ def get_marking_scheme(self):
+ return self.section_marking_scheme
+
+ def get_section_explanation(self):
+ answer_type = self.answer_type
+ if answer_type in ["standard", "multiple-correct"]:
+ return self.section_marking_scheme.section_key
+ elif answer_type == "multiple-correct-weighted":
+ return f"Custom: {self.marking}"
+
+ def get_verdict_marking(self, marked_answer):
+ answer_type = self.answer_type
+ question_verdict = "incorrect"
+ if answer_type == "standard":
+ question_verdict = self.get_standard_verdict(marked_answer)
+ elif answer_type == "multiple-correct":
+ question_verdict = self.get_multiple_correct_verdict(marked_answer)
+ elif answer_type == "multiple-correct-weighted":
+ question_verdict = self.get_multiple_correct_weighted_verdict(marked_answer)
+ return question_verdict, self.marking[question_verdict]
+
+ def get_standard_verdict(self, marked_answer):
+ allowed_answer = self.answer_item
+ if marked_answer == self.empty_val:
+ return "unmarked"
+ elif marked_answer == allowed_answer:
+ return "correct"
+ else:
+ return "incorrect"
+
+ def get_multiple_correct_verdict(self, marked_answer):
+ allowed_answers = self.answer_item
+ if marked_answer == self.empty_val:
+ return "unmarked"
+ elif marked_answer in allowed_answers:
+ return f"correct-{marked_answer}"
+ else:
+ return "incorrect"
+
+ def get_multiple_correct_weighted_verdict(self, marked_answer):
+ allowed_answers = [
+ allowed_answer for allowed_answer, _answer_score in self.answer_item
+ ]
+ if marked_answer == self.empty_val:
+ return "unmarked"
+ elif marked_answer in allowed_answers:
+ return f"correct-{marked_answer}"
+ else:
+ return "incorrect"
+
+ def __str__(self):
+ return f"{self.answer_item}"
+
+
+class SectionMarkingScheme:
+ def __init__(self, section_key, section_scheme, empty_val):
+ # TODO: get local empty_val from qblock
+ self.empty_val = empty_val
+ self.section_key = section_key
+ # DEFAULT marking scheme follows a shorthand
+ if section_key == DEFAULT_SECTION_KEY:
+ self.questions = None
+ self.marking = self.parse_scheme_marking(section_scheme)
+ else:
+ self.questions = parse_fields(section_key, section_scheme["questions"])
+ self.marking = self.parse_scheme_marking(section_scheme["marking"])
+
+ def __str__(self):
+ return self.section_key
+
+ def parse_scheme_marking(self, marking):
+ parsed_marking = {}
+ for verdict_type in MARKING_VERDICT_TYPES:
+ verdict_marking = parse_float_or_fraction(marking[verdict_type])
+ if (
+ verdict_marking > 0
+ and verdict_type == "incorrect"
+ and not self.section_key.startswith(BONUS_SECTION_PREFIX)
+ ):
+ logger.warning(
+ f"Found positive marks({round(verdict_marking, 2)}) for incorrect answer in the schema '{self.section_key}'. For Bonus sections, add a prefix 'BONUS_' to them."
+ )
+ parsed_marking[verdict_type] = verdict_marking
+
+ return parsed_marking
+
+ def match_answer(self, marked_answer, answer_matcher):
+ question_verdict, verdict_marking = answer_matcher.get_verdict_marking(
+ marked_answer
+ )
+
+ return verdict_marking, question_verdict
+
+
+class EvaluationConfig:
+ """Note: this instance will be reused for multiple omr sheets"""
+
+ def __init__(self, curr_dir, evaluation_path, template, tuning_config):
+ self.path = evaluation_path
+ evaluation_json = open_evaluation_with_validation(evaluation_path)
+ options, marking_schemes, source_type = map(
+ evaluation_json.get, ["options", "marking_schemes", "source_type"]
+ )
+ self.should_explain_scoring = options.get("should_explain_scoring", False)
+ self.has_non_default_section = False
+ self.exclude_files = []
+ self.enable_evaluation_table_to_csv = options.get(
+ "enable_evaluation_table_to_csv", False
+ )
+
+ if source_type == "csv":
+ csv_path = curr_dir.joinpath(options["answer_key_csv_path"])
+ if not os.path.exists(csv_path):
+ logger.warning(f"Answer key csv does not exist at: '{csv_path}'.")
+
+ answer_key_image_path = options.get("answer_key_image_path", None)
+ if os.path.exists(csv_path):
+ # TODO: CSV parsing/validation for each row with a (qNo, ) pair
+ answer_key = pd.read_csv(
+ csv_path,
+ header=None,
+ names=["question", "answer"],
+ converters={"question": str, "answer": self.parse_answer_column},
+ )
+
+ self.questions_in_order = answer_key["question"].to_list()
+ answers_in_order = answer_key["answer"].to_list()
+ elif not answer_key_image_path:
+ raise Exception(f"Answer key csv not found at '{csv_path}'")
+ else:
+ image_path = str(curr_dir.joinpath(answer_key_image_path))
+ if not os.path.exists(image_path):
+ raise Exception(f"Answer key image not found at '{image_path}'")
+
+ # self.exclude_files.append(image_path)
+
+ logger.debug(
+ f"Attempting to generate answer key from image: '{image_path}'"
+ )
+ # TODO: use a common function for below changes?
+ in_omr = cv2.imread(image_path, cv2.IMREAD_GRAYSCALE)
+ in_omr = template.image_instance_ops.apply_preprocessors(
+ image_path, in_omr, template
+ )
+ if in_omr is None:
+ raise Exception(
+ f"Could not read answer key from image {image_path}"
+ )
+ (
+ response_dict,
+ _final_marked,
+ _multi_marked,
+ _multi_roll,
+ ) = template.image_instance_ops.read_omr_response(
+ template,
+ image=in_omr,
+ name=image_path,
+ save_dir=None,
+ )
+ omr_response = get_concatenated_response(response_dict, template)
+
+ empty_val = template.global_empty_val
+ empty_answer_regex = (
+ rf"{re.escape(empty_val)}+" if empty_val != "" else r"^$"
+ )
+
+ if "questions_in_order" in options:
+ self.questions_in_order = self.parse_questions_in_order(
+ options["questions_in_order"]
+ )
+ empty_answered_questions = [
+ question
+ for question in self.questions_in_order
+ if re.search(empty_answer_regex, omr_response[question])
+ ]
+ if len(empty_answered_questions) > 0:
+ logger.error(
+ f"Found empty answers for questions: {empty_answered_questions}, empty value used: '{empty_val}'"
+ )
+ raise Exception(
+ f"Found empty answers in file '{image_path}'. Please check your template again in the --setLayout mode."
+ )
+ else:
+ logger.warning(
+ f"questions_in_order not provided, proceeding to use non-empty values as answer key"
+ )
+ self.questions_in_order = sorted(
+ question
+ for (question, answer) in omr_response.items()
+ if not re.search(empty_answer_regex, answer)
+ )
+ answers_in_order = [
+ omr_response[question] for question in self.questions_in_order
+ ]
+ # TODO: save the CSV
+ else:
+ self.questions_in_order = self.parse_questions_in_order(
+ options["questions_in_order"]
+ )
+ answers_in_order = options["answers_in_order"]
+
+ self.validate_questions(answers_in_order)
+
+ self.section_marking_schemes, self.question_to_scheme = {}, {}
+ for section_key, section_scheme in marking_schemes.items():
+ section_marking_scheme = SectionMarkingScheme(
+ section_key, section_scheme, template.global_empty_val
+ )
+ if section_key != DEFAULT_SECTION_KEY:
+ self.section_marking_schemes[section_key] = section_marking_scheme
+ for q in section_marking_scheme.questions:
+ # TODO: check the answer key for custom scheme here?
+ self.question_to_scheme[q] = section_marking_scheme
+ self.has_non_default_section = True
+ else:
+ self.default_marking_scheme = section_marking_scheme
+
+ self.validate_marking_schemes()
+
+ self.question_to_answer_matcher = self.parse_answers_and_map_questions(
+ answers_in_order
+ )
+ self.validate_answers(answers_in_order, tuning_config)
+
+ def __str__(self):
+ return str(self.path)
+
+ # Externally called methods have higher abstraction level.
+ def prepare_and_validate_omr_response(self, omr_response):
+ self.reset_explanation_table()
+
+ omr_response_questions = set(omr_response.keys())
+ all_questions = set(self.questions_in_order)
+ missing_questions = sorted(all_questions.difference(omr_response_questions))
+ if len(missing_questions) > 0:
+ logger.critical(f"Missing OMR response for: {missing_questions}")
+ raise Exception(
+ f"Some questions are missing in the OMR response for the given answer key"
+ )
+
+ prefixed_omr_response_questions = set(
+ [k for k in omr_response.keys() if k.startswith("q")]
+ )
+ missing_prefixed_questions = sorted(
+ prefixed_omr_response_questions.difference(all_questions)
+ )
+ if len(missing_prefixed_questions) > 0:
+ logger.warning(
+ f"No answer given for potential questions in OMR response: {missing_prefixed_questions}"
+ )
+
+ def match_answer_for_question(self, current_score, question, marked_answer):
+ answer_matcher = self.question_to_answer_matcher[question]
+ question_verdict, delta = answer_matcher.get_verdict_marking(marked_answer)
+ self.conditionally_add_explanation(
+ answer_matcher,
+ delta,
+ marked_answer,
+ question_verdict,
+ question,
+ current_score,
+ )
+ return delta
+
+ def conditionally_print_explanation(self):
+ if self.should_explain_scoring:
+ console.print(self.explanation_table, justify="center")
+
+ # Explanation Table to CSV
+ def conditionally_save_explanation_csv(self, file_path, evaluation_output_dir):
+ if self.enable_evaluation_table_to_csv:
+ data = {col.header: col._cells for col in self.explanation_table.columns}
+
+ output_path = os.path.join(
+ evaluation_output_dir,
+ f"{file_path.stem}_evaluation.csv",
+ )
+
+ pd.DataFrame(data, dtype=str).to_csv(
+ output_path,
+ mode="a",
+ quoting=QUOTE_NONNUMERIC,
+ index=False,
+ )
+
+ def get_should_explain_scoring(self):
+ return self.should_explain_scoring
+
+ def get_exclude_files(self):
+ return self.exclude_files
+
+ @staticmethod
+ def parse_answer_column(answer_column):
+ if answer_column[0] == "[":
+ # multiple-correct-weighted or multiple-correct
+ parsed_answer = ast.literal_eval(answer_column)
+ elif "," in answer_column:
+ # multiple-correct
+ parsed_answer = answer_column.split(",")
+ else:
+ # single-correct
+ parsed_answer = answer_column
+ return parsed_answer
+
+ def parse_questions_in_order(self, questions_in_order):
+ return parse_fields("questions_in_order", questions_in_order)
+
+ def validate_answers(self, answers_in_order, tuning_config):
+ answer_matcher_map = self.question_to_answer_matcher
+ if tuning_config.outputs.filter_out_multimarked_files:
+ multi_marked_answer = False
+ for question, answer_item in zip(self.questions_in_order, answers_in_order):
+ answer_type = answer_matcher_map[question].answer_type
+ if answer_type == "standard":
+ if len(answer_item) > 1:
+ multi_marked_answer = True
+ if answer_type == "multiple-correct":
+ for single_answer in answer_item:
+ if len(single_answer) > 1:
+ multi_marked_answer = True
+ break
+ if answer_type == "multiple-correct-weighted":
+ for single_answer, _answer_score in answer_item:
+ if len(single_answer) > 1:
+ multi_marked_answer = True
+
+ if multi_marked_answer:
+ raise Exception(
+ f"Provided answer key contains multiple correct answer(s), but config.filter_out_multimarked_files is True. Scoring will get skipped."
+ )
+
+ def validate_questions(self, answers_in_order):
+ questions_in_order = self.questions_in_order
+ len_questions_in_order, len_answers_in_order = len(questions_in_order), len(
+ answers_in_order
+ )
+ if len_questions_in_order != len_answers_in_order:
+ logger.critical(
+ f"questions_in_order({len_questions_in_order}): {questions_in_order}\nanswers_in_order({len_answers_in_order}): {answers_in_order}"
+ )
+ raise Exception(
+ f"Unequal lengths for questions_in_order and answers_in_order ({len_questions_in_order} != {len_answers_in_order})"
+ )
+
+ def validate_marking_schemes(self):
+ section_marking_schemes = self.section_marking_schemes
+ section_questions = set()
+ for section_key, section_scheme in section_marking_schemes.items():
+ if section_key == DEFAULT_SECTION_KEY:
+ continue
+ current_set = set(section_scheme.questions)
+ if not section_questions.isdisjoint(current_set):
+ raise Exception(
+ f"Section '{section_key}' has overlapping question(s) with other sections"
+ )
+ section_questions = section_questions.union(current_set)
+
+ all_questions = set(self.questions_in_order)
+ missing_questions = sorted(section_questions.difference(all_questions))
+ if len(missing_questions) > 0:
+ logger.critical(f"Missing answer key for: {missing_questions}")
+ raise Exception(
+ f"Some questions are missing in the answer key for the given marking scheme"
+ )
+
+ def parse_answers_and_map_questions(self, answers_in_order):
+ question_to_answer_matcher = {}
+ for question, answer_item in zip(self.questions_in_order, answers_in_order):
+ section_marking_scheme = self.get_marking_scheme_for_question(question)
+ answer_matcher = AnswerMatcher(answer_item, section_marking_scheme)
+ question_to_answer_matcher[question] = answer_matcher
+ if (
+ answer_matcher.answer_type == "multiple-correct-weighted"
+ and section_marking_scheme.section_key != DEFAULT_SECTION_KEY
+ ):
+ logger.warning(
+ f"The custom scheme '{section_marking_scheme}' will not apply to question '{question}' as it will use the given answer weights f{answer_item}"
+ )
+ return question_to_answer_matcher
+
+ # Then unfolding lower abstraction levels
+ def reset_explanation_table(self):
+ self.explanation_table = None
+ self.prepare_explanation_table()
+
+ def prepare_explanation_table(self):
+ # TODO: provide a way to export this as csv/pdf
+ if not self.should_explain_scoring:
+ return
+ table = Table(title="Evaluation Explanation Table", show_lines=True)
+ table.add_column("Question")
+ table.add_column("Marked")
+ table.add_column("Answer(s)")
+ table.add_column("Verdict")
+ table.add_column("Delta")
+ table.add_column("Score")
+ # TODO: Add max and min score in explanation (row-wise and total)
+ if self.has_non_default_section:
+ table.add_column("Section")
+ self.explanation_table = table
+
+ def get_marking_scheme_for_question(self, question):
+ return self.question_to_scheme.get(question, self.default_marking_scheme)
+
+ def conditionally_add_explanation(
+ self,
+ answer_matcher,
+ delta,
+ marked_answer,
+ question_verdict,
+ question,
+ current_score,
+ ):
+ if self.should_explain_scoring:
+ next_score = current_score + delta
+ # Conditionally add cells
+ row = [
+ item
+ for item in [
+ question,
+ marked_answer,
+ str(answer_matcher),
+ str.title(question_verdict),
+ str(round(delta, 2)),
+ str(round(next_score, 2)),
+ (
+ answer_matcher.get_section_explanation()
+ if self.has_non_default_section
+ else None
+ ),
+ ]
+ if item is not None
+ ]
+ self.explanation_table.add_row(*row)
+
+
+def evaluate_concatenated_response(
+ concatenated_response, evaluation_config, file_path, evaluation_output_dir
+):
+ evaluation_config.prepare_and_validate_omr_response(concatenated_response)
+ current_score = 0.0
+ for question in evaluation_config.questions_in_order:
+ marked_answer = concatenated_response[question]
+ delta = evaluation_config.match_answer_for_question(
+ current_score, question, marked_answer
+ )
+ current_score += delta
+
+ evaluation_config.conditionally_print_explanation()
+ evaluation_config.conditionally_save_explanation_csv(file_path, evaluation_output_dir)
+
+ return current_score
diff --git a/src/logger.py b/src/logger.py
new file mode 100644
index 00000000..27f44b44
--- /dev/null
+++ b/src/logger.py
@@ -0,0 +1,68 @@
+import logging
+from typing import Union
+
+from rich.console import Console
+from rich.logging import RichHandler
+
+FORMAT = "%(message)s"
+
+# TODO: set logging level from config.json dynamically
+logging.basicConfig(
+ level=logging.INFO,
+ format="%(message)s",
+ datefmt="[%X]",
+ handlers=[RichHandler(rich_tracebacks=True)],
+)
+
+
+class Logger:
+ def __init__(
+ self,
+ name,
+ level: Union[int, str] = logging.NOTSET,
+ message_format="%(message)s",
+ date_format="[%X]",
+ ):
+ self.log = logging.getLogger(name)
+ self.log.setLevel(level)
+ self.log.__format__ = message_format
+ self.log.__date_format__ = date_format
+
+ def debug(self, *msg: object, sep=" ", end="\n") -> None:
+ return self.logutil("debug", *msg, sep=sep)
+
+ def info(self, *msg: object, sep=" ", end="\n") -> None:
+ return self.logutil("info", *msg, sep=sep)
+
+ def warning(self, *msg: object, sep=" ", end="\n") -> None:
+ return self.logutil("warning", *msg, sep=sep)
+
+ def error(self, *msg: object, sep=" ", end="\n") -> None:
+ return self.logutil("error", *msg, sep=sep)
+
+ def critical(self, *msg: object, sep=" ", end="\n") -> None:
+ return self.logutil("critical", *msg, sep=sep)
+
+ def stringify(func):
+ def inner(self, method_type: str, *msg: object, sep=" "):
+ nmsg = []
+ for v in msg:
+ if not isinstance(v, str):
+ v = str(v)
+ nmsg.append(v)
+ return func(self, method_type, *nmsg, sep=sep)
+
+ return inner
+
+ # set stack level to 3 so that the caller of this function is logged, not this function itself.
+ # stack-frame - self.log.debug - logutil - stringify - log method - caller
+ @stringify
+ def logutil(self, method_type: str, *msg: object, sep=" ") -> None:
+ func = getattr(self.log, method_type, None)
+ if not func:
+ raise AttributeError(f"Logger has no method {method_type}")
+ return func(sep.join(msg), stacklevel=4)
+
+
+logger = Logger(__name__)
+console = Console()
diff --git a/src/processors/CropOnMarkers.py b/src/processors/CropOnMarkers.py
new file mode 100644
index 00000000..4867e038
--- /dev/null
+++ b/src/processors/CropOnMarkers.py
@@ -0,0 +1,233 @@
+import os
+
+import cv2
+import numpy as np
+
+from src.logger import logger
+from src.processors.interfaces.ImagePreprocessor import ImagePreprocessor
+from src.utils.image import ImageUtils
+from src.utils.interaction import InteractionUtils
+
+
+class CropOnMarkers(ImagePreprocessor):
+ def __init__(self, *args, **kwargs):
+ super().__init__(*args, **kwargs)
+ config = self.tuning_config
+ marker_ops = self.options
+ self.threshold_circles = []
+ # img_utils = ImageUtils()
+
+ # options with defaults
+ self.marker_path = os.path.join(
+ self.relative_dir, marker_ops.get("relativePath", "omr_marker.jpg")
+ )
+ self.min_matching_threshold = marker_ops.get("min_matching_threshold", 0.3)
+ self.max_matching_variation = marker_ops.get("max_matching_variation", 0.41)
+ self.marker_rescale_range = tuple(
+ int(r) for r in marker_ops.get("marker_rescale_range", (35, 100))
+ )
+ self.marker_rescale_steps = int(marker_ops.get("marker_rescale_steps", 10))
+ self.apply_erode_subtract = marker_ops.get("apply_erode_subtract", True)
+ self.marker = self.load_marker(marker_ops, config)
+
+ def __str__(self):
+ return self.marker_path
+
+ def exclude_files(self):
+ return [self.marker_path]
+
+ def apply_filter(self, image, file_path):
+ config = self.tuning_config
+ image_instance_ops = self.image_instance_ops
+ image_eroded_sub = ImageUtils.normalize_util(
+ image
+ if self.apply_erode_subtract
+ else (image - cv2.erode(image, kernel=np.ones((5, 5)), iterations=5))
+ )
+ # Quads on warped image
+ quads = {}
+ h1, w1 = image_eroded_sub.shape[:2]
+ midh, midw = h1 // 3, w1 // 2
+ origins = [[0, 0], [midw, 0], [0, midh], [midw, midh]]
+ quads[0] = image_eroded_sub[0:midh, 0:midw]
+ quads[1] = image_eroded_sub[0:midh, midw:w1]
+ quads[2] = image_eroded_sub[midh:h1, 0:midw]
+ quads[3] = image_eroded_sub[midh:h1, midw:w1]
+
+ # Draw Quadlines
+ image_eroded_sub[:, midw : midw + 2] = 255
+ image_eroded_sub[midh : midh + 2, :] = 255
+
+ best_scale, all_max_t = self.getBestMatch(image_eroded_sub)
+ if best_scale is None:
+ if config.outputs.show_image_level >= 1:
+ InteractionUtils.show("Quads", image_eroded_sub, config=config)
+ return None
+
+ optimal_marker = ImageUtils.resize_util_h(
+ self.marker, u_height=int(self.marker.shape[0] * best_scale)
+ )
+ _h, w = optimal_marker.shape[:2]
+ centres = []
+ sum_t, max_t = 0, 0
+ quarter_match_log = "Matching Marker: "
+ for k in range(0, 4):
+ res = cv2.matchTemplate(quads[k], optimal_marker, cv2.TM_CCOEFF_NORMED)
+ max_t = res.max()
+ quarter_match_log += f"Quarter{str(k + 1)}: {str(round(max_t, 3))}\t"
+ if (
+ max_t < self.min_matching_threshold
+ or abs(all_max_t - max_t) >= self.max_matching_variation
+ ):
+ logger.error(
+ file_path,
+ "\nError: No circle found in Quad",
+ k + 1,
+ "\n\t min_matching_threshold",
+ self.min_matching_threshold,
+ "\t max_matching_variation",
+ self.max_matching_variation,
+ "\t max_t",
+ max_t,
+ "\t all_max_t",
+ all_max_t,
+ )
+ if config.outputs.show_image_level >= 1:
+ InteractionUtils.show(
+ f"No markers: {file_path}",
+ image_eroded_sub,
+ 0,
+ config=config,
+ )
+ InteractionUtils.show(
+ f"res_Q{str(k + 1)} ({str(max_t)})",
+ res,
+ 1,
+ config=config,
+ )
+ return None
+
+ pt = np.argwhere(res == max_t)[0]
+ pt = [pt[1], pt[0]]
+ pt[0] += origins[k][0]
+ pt[1] += origins[k][1]
+ # print(">>",pt)
+ image = cv2.rectangle(
+ image, tuple(pt), (pt[0] + w, pt[1] + _h), (150, 150, 150), 2
+ )
+ # display:
+ image_eroded_sub = cv2.rectangle(
+ image_eroded_sub,
+ tuple(pt),
+ (pt[0] + w, pt[1] + _h),
+ (50, 50, 50) if self.apply_erode_subtract else (155, 155, 155),
+ 4,
+ )
+ centres.append([pt[0] + w / 2, pt[1] + _h / 2])
+ sum_t += max_t
+
+ logger.info(quarter_match_log)
+ logger.info(f"Optimal Scale: {best_scale}")
+ # analysis data
+ self.threshold_circles.append(sum_t / 4)
+
+ image = ImageUtils.four_point_transform(image, np.array(centres))
+ # appendSaveImg(1,image_eroded_sub)
+ # appendSaveImg(1,image_norm)
+
+ image_instance_ops.append_save_img(2, image_eroded_sub)
+ # Debugging image -
+ # res = cv2.matchTemplate(image_eroded_sub,optimal_marker,cv2.TM_CCOEFF_NORMED)
+ # res[ : , midw:midw+2] = 255
+ # res[ midh:midh+2, : ] = 255
+ # show("Markers Matching",res)
+ if config.outputs.show_image_level >= 2 and config.outputs.show_image_level < 4:
+ image_eroded_sub = ImageUtils.resize_util_h(
+ image_eroded_sub, image.shape[0]
+ )
+ image_eroded_sub[:, -5:] = 0
+ h_stack = np.hstack((image_eroded_sub, image))
+ InteractionUtils.show(
+ f"Warped: {file_path}",
+ ImageUtils.resize_util(
+ h_stack, int(config.dimensions.display_width * 1.6)
+ ),
+ 0,
+ 0,
+ [0, 0],
+ config=config,
+ )
+ # iterations : Tuned to 2.
+ # image_eroded_sub = image_norm - cv2.erode(image_norm, kernel=np.ones((5,5)),iterations=2)
+ return image
+
+ def load_marker(self, marker_ops, config):
+ if not os.path.exists(self.marker_path):
+ logger.error(
+ "Marker not found at path provided in template:",
+ self.marker_path,
+ )
+ exit(31)
+
+ marker = cv2.imread(self.marker_path, cv2.IMREAD_GRAYSCALE)
+
+ if "sheetToMarkerWidthRatio" in marker_ops:
+ marker = ImageUtils.resize_util(
+ marker,
+ config.dimensions.processing_width
+ / int(marker_ops["sheetToMarkerWidthRatio"]),
+ )
+ marker = cv2.GaussianBlur(marker, (5, 5), 0)
+ marker = cv2.normalize(
+ marker, None, alpha=0, beta=255, norm_type=cv2.NORM_MINMAX
+ )
+
+ if self.apply_erode_subtract:
+ marker -= cv2.erode(marker, kernel=np.ones((5, 5)), iterations=5)
+
+ return marker
+
+ # Resizing the marker within scaleRange at rate of descent_per_step to
+ # find the best match.
+ def getBestMatch(self, image_eroded_sub):
+ config = self.tuning_config
+ descent_per_step = (
+ self.marker_rescale_range[1] - self.marker_rescale_range[0]
+ ) // self.marker_rescale_steps
+ _h, _w = self.marker.shape[:2]
+ res, best_scale = None, None
+ all_max_t = 0
+
+ for r0 in np.arange(
+ self.marker_rescale_range[1],
+ self.marker_rescale_range[0],
+ -1 * descent_per_step,
+ ): # reverse order
+ s = float(r0 * 1 / 100)
+ if s == 0.0:
+ continue
+ rescaled_marker = ImageUtils.resize_util_h(
+ self.marker, u_height=int(_h * s)
+ )
+ # res is the black image with white dots
+ res = cv2.matchTemplate(
+ image_eroded_sub, rescaled_marker, cv2.TM_CCOEFF_NORMED
+ )
+
+ max_t = res.max()
+ if all_max_t < max_t:
+ # print('Scale: '+str(s)+', Circle Match: '+str(round(max_t*100,2))+'%')
+ best_scale, all_max_t = s, max_t
+
+ if all_max_t < self.min_matching_threshold:
+ logger.warning(
+ "\tTemplate matching too low! Consider rechecking preProcessors applied before this."
+ )
+ if config.outputs.show_image_level >= 1:
+ InteractionUtils.show("res", res, 1, 0, config=config)
+
+ if best_scale is None:
+ logger.warning(
+ "No matchings for given scaleRange:", self.marker_rescale_range
+ )
+ return best_scale, all_max_t
diff --git a/src/processors/CropPage.py b/src/processors/CropPage.py
new file mode 100644
index 00000000..25629e41
--- /dev/null
+++ b/src/processors/CropPage.py
@@ -0,0 +1,112 @@
+"""
+https://www.pyimagesearch.com/2015/04/06/zero-parameter-automatic-canny-edge-detection-with-python-and-opencv/
+"""
+import cv2
+import numpy as np
+
+from src.logger import logger
+from src.processors.interfaces.ImagePreprocessor import ImagePreprocessor
+from src.utils.image import ImageUtils
+from src.utils.interaction import InteractionUtils
+
+MIN_PAGE_AREA = 80000
+
+
+def normalize(image):
+ return cv2.normalize(image, 0, 255, norm_type=cv2.NORM_MINMAX)
+
+
+def check_max_cosine(approx):
+ # assumes 4 pts present
+ max_cosine = 0
+ min_cosine = 1.5
+ for i in range(2, 5):
+ cosine = abs(angle(approx[i % 4], approx[i - 2], approx[i - 1]))
+ max_cosine = max(cosine, max_cosine)
+ min_cosine = min(cosine, min_cosine)
+
+ if max_cosine >= 0.35:
+ logger.warning("Quadrilateral is not a rectangle.")
+ return False
+ return True
+
+
+def validate_rect(approx):
+ return len(approx) == 4 and check_max_cosine(approx.reshape(4, 2))
+
+
+def angle(p_1, p_2, p_0):
+ dx1 = float(p_1[0] - p_0[0])
+ dy1 = float(p_1[1] - p_0[1])
+ dx2 = float(p_2[0] - p_0[0])
+ dy2 = float(p_2[1] - p_0[1])
+ return (dx1 * dx2 + dy1 * dy2) / np.sqrt(
+ (dx1 * dx1 + dy1 * dy1) * (dx2 * dx2 + dy2 * dy2) + 1e-10
+ )
+
+
+class CropPage(ImagePreprocessor):
+ def __init__(self, *args, **kwargs):
+ super().__init__(*args, **kwargs)
+ cropping_ops = self.options
+ self.morph_kernel = tuple(
+ int(x) for x in cropping_ops.get("morphKernel", [10, 10])
+ )
+
+ def apply_filter(self, image, file_path):
+ image = normalize(cv2.GaussianBlur(image, (3, 3), 0))
+
+ # Resize should be done with another preprocessor is needed
+ sheet = self.find_page(image, file_path)
+ if len(sheet) == 0:
+ logger.error(
+ f"\tError: Paper boundary not found for: '{file_path}'\nHave you accidentally included CropPage preprocessor?"
+ )
+ return None
+
+ logger.info(f"Found page corners: \t {sheet.tolist()}")
+
+ # Warp layer 1
+ image = ImageUtils.four_point_transform(image, sheet)
+
+ # Return preprocessed image
+ return image
+
+ def find_page(self, image, file_path):
+ config = self.tuning_config
+
+ image = normalize(image)
+
+ _ret, image = cv2.threshold(image, 200, 255, cv2.THRESH_TRUNC)
+ image = normalize(image)
+
+ kernel = cv2.getStructuringElement(cv2.MORPH_RECT, self.morph_kernel)
+
+ # Close the small holes, i.e. Complete the edges on canny image
+ closed = cv2.morphologyEx(image, cv2.MORPH_CLOSE, kernel)
+
+ edge = cv2.Canny(closed, 185, 55)
+
+ if config.outputs.show_image_level >= 5:
+ InteractionUtils.show("edge", edge, config=config)
+
+ # findContours returns outer boundaries in CW and inner ones, ACW.
+ cnts = ImageUtils.grab_contours(
+ cv2.findContours(edge, cv2.RETR_LIST, cv2.CHAIN_APPROX_SIMPLE)
+ )
+ # convexHull to resolve disordered curves due to noise
+ cnts = [cv2.convexHull(c) for c in cnts]
+ cnts = sorted(cnts, key=cv2.contourArea, reverse=True)[:5]
+ sheet = []
+ for c in cnts:
+ if cv2.contourArea(c) < MIN_PAGE_AREA:
+ continue
+ peri = cv2.arcLength(c, True)
+ approx = cv2.approxPolyDP(c, epsilon=0.025 * peri, closed=True)
+ if validate_rect(approx):
+ sheet = np.reshape(approx, (4, -1))
+ cv2.drawContours(image, [approx], -1, (0, 255, 0), 2)
+ cv2.drawContours(edge, [approx], -1, (255, 255, 255), 10)
+ break
+
+ return sheet
diff --git a/src/processors/FeatureBasedAlignment.py b/src/processors/FeatureBasedAlignment.py
new file mode 100644
index 00000000..be872924
--- /dev/null
+++ b/src/processors/FeatureBasedAlignment.py
@@ -0,0 +1,94 @@
+"""
+Image based feature alignment
+Credits: https://www.learnopencv.com/image-alignment-feature-based-using-opencv-c-python/
+"""
+import cv2
+import numpy as np
+
+from src.processors.interfaces.ImagePreprocessor import ImagePreprocessor
+from src.utils.image import ImageUtils
+from src.utils.interaction import InteractionUtils
+
+
+class FeatureBasedAlignment(ImagePreprocessor):
+ def __init__(self, *args, **kwargs):
+ super().__init__(*args, **kwargs)
+ options = self.options
+ config = self.tuning_config
+
+ # process reference image
+ self.ref_path = self.relative_dir.joinpath(options["reference"])
+ ref_img = cv2.imread(str(self.ref_path), cv2.IMREAD_GRAYSCALE)
+ self.ref_img = ImageUtils.resize_util(
+ ref_img,
+ config.dimensions.processing_width,
+ config.dimensions.processing_height,
+ )
+ # get options with defaults
+ self.max_features = int(options.get("maxFeatures", 500))
+ self.good_match_percent = options.get("goodMatchPercent", 0.15)
+ self.transform_2_d = options.get("2d", False)
+ # Extract keypoints and description of source image
+ self.orb = cv2.ORB_create(self.max_features)
+ self.to_keypoints, self.to_descriptors = self.orb.detectAndCompute(
+ self.ref_img, None
+ )
+
+ def __str__(self):
+ return self.ref_path.name
+
+ def exclude_files(self):
+ return [self.ref_path]
+
+ def apply_filter(self, image, _file_path):
+ config = self.tuning_config
+ # Convert images to grayscale
+ # im1Gray = cv2.cvtColor(im1, cv2.COLOR_BGR2GRAY)
+ # im2Gray = cv2.cvtColor(im2, cv2.COLOR_BGR2GRAY)
+
+ image = cv2.normalize(image, 0, 255, norm_type=cv2.NORM_MINMAX)
+
+ # Detect ORB features and compute descriptors.
+ from_keypoints, from_descriptors = self.orb.detectAndCompute(image, None)
+
+ # Match features.
+ matcher = cv2.DescriptorMatcher_create(
+ cv2.DESCRIPTOR_MATCHER_BRUTEFORCE_HAMMING
+ )
+
+ # create BFMatcher object (alternate matcher)
+ # matcher = cv2.BFMatcher(cv2.NORM_HAMMING, crossCheck=True)
+
+ matches = np.array(matcher.match(from_descriptors, self.to_descriptors, None))
+
+ # Sort matches by score
+ matches = sorted(matches, key=lambda x: x.distance, reverse=False)
+
+ # Remove not so good matches
+ num_good_matches = int(len(matches) * self.good_match_percent)
+ matches = matches[:num_good_matches]
+
+ # Draw top matches
+ if config.outputs.show_image_level > 2:
+ im_matches = cv2.drawMatches(
+ image, from_keypoints, self.ref_img, self.to_keypoints, matches, None
+ )
+ InteractionUtils.show("Aligning", im_matches, resize=True, config=config)
+
+ # Extract location of good matches
+ points1 = np.zeros((len(matches), 2), dtype=np.float32)
+ points2 = np.zeros((len(matches), 2), dtype=np.float32)
+
+ for i, match in enumerate(matches):
+ points1[i, :] = from_keypoints[match.queryIdx].pt
+ points2[i, :] = self.to_keypoints[match.trainIdx].pt
+
+ # Find homography
+ height, width = self.ref_img.shape
+ if self.transform_2_d:
+ m, _inliers = cv2.estimateAffine2D(points1, points2)
+ return cv2.warpAffine(image, m, (width, height))
+
+ # Use homography
+ h, _mask = cv2.findHomography(points1, points2, cv2.RANSAC)
+ return cv2.warpPerspective(image, h, (width, height))
diff --git a/src/processors/builtins.py b/src/processors/builtins.py
new file mode 100644
index 00000000..faa6288f
--- /dev/null
+++ b/src/processors/builtins.py
@@ -0,0 +1,54 @@
+import cv2
+import numpy as np
+
+from src.processors.interfaces.ImagePreprocessor import ImagePreprocessor
+
+
+class Levels(ImagePreprocessor):
+ def __init__(self, *args, **kwargs):
+ super().__init__(*args, **kwargs)
+ options = self.options
+
+ def output_level(value, low, high, gamma):
+ if value <= low:
+ return 0
+ if value >= high:
+ return 255
+ inv_gamma = 1.0 / gamma
+ return (((value - low) / (high - low)) ** inv_gamma) * 255
+
+ self.gamma = np.array(
+ [
+ output_level(
+ i,
+ int(255 * options.get("low", 0)),
+ int(255 * options.get("high", 1)),
+ options.get("gamma", 1.0),
+ )
+ for i in np.arange(0, 256)
+ ]
+ ).astype("uint8")
+
+ def apply_filter(self, image, _file_path):
+ return cv2.LUT(image, self.gamma)
+
+
+class MedianBlur(ImagePreprocessor):
+ def __init__(self, *args, **kwargs):
+ super().__init__(*args, **kwargs)
+ options = self.options
+ self.kSize = int(options.get("kSize", 5))
+
+ def apply_filter(self, image, _file_path):
+ return cv2.medianBlur(image, self.kSize)
+
+
+class GaussianBlur(ImagePreprocessor):
+ def __init__(self, *args, **kwargs):
+ super().__init__(*args, **kwargs)
+ options = self.options
+ self.kSize = tuple(int(x) for x in options.get("kSize", (3, 3)))
+ self.sigmaX = int(options.get("sigmaX", 0))
+
+ def apply_filter(self, image, _file_path):
+ return cv2.GaussianBlur(image, self.kSize, self.sigmaX)
diff --git a/src/processors/interfaces/ImagePreprocessor.py b/src/processors/interfaces/ImagePreprocessor.py
new file mode 100644
index 00000000..a5cf09a4
--- /dev/null
+++ b/src/processors/interfaces/ImagePreprocessor.py
@@ -0,0 +1,18 @@
+# Use all imports relative to root directory
+from src.processors.manager import Processor
+
+
+class ImagePreprocessor(Processor):
+ """Base class for an extension that applies some preprocessing to the input image"""
+
+ def __init__(self, *args, **kwargs):
+ super().__init__(*args, **kwargs)
+
+ def apply_filter(self, image, filename):
+ """Apply filter to the image and returns modified image"""
+ raise NotImplementedError
+
+ @staticmethod
+ def exclude_files():
+ """Returns a list of file paths that should be excluded from processing"""
+ return []
diff --git a/src/processors/manager.py b/src/processors/manager.py
new file mode 100644
index 00000000..c41e0713
--- /dev/null
+++ b/src/processors/manager.py
@@ -0,0 +1,80 @@
+"""
+Processor/Extension framework
+Adapated from https://github.com/gdiepen/python_processor_example
+"""
+import inspect
+import pkgutil
+
+from src.logger import logger
+
+
+class Processor:
+ """Base class that each processor must inherit from."""
+
+ def __init__(
+ self,
+ options=None,
+ relative_dir=None,
+ image_instance_ops=None,
+ ):
+ self.options = options
+ self.relative_dir = relative_dir
+ self.image_instance_ops = image_instance_ops
+ self.tuning_config = image_instance_ops.tuning_config
+ self.description = "UNKNOWN"
+
+
+class ProcessorManager:
+ """Upon creation, this class will read the processors package for modules
+ that contain a class definition that is inheriting from the Processor class
+ """
+
+ def __init__(self, processors_dir="src.processors"):
+ """Constructor that initiates the reading of all available processors
+ when an instance of the ProcessorCollection object is created
+ """
+ self.processors_dir = processors_dir
+ self.reload_processors()
+
+ @staticmethod
+ def get_name_filter(processor_name):
+ def filter_function(member):
+ return inspect.isclass(member) and member.__module__ == processor_name
+
+ return filter_function
+
+ def reload_processors(self):
+ """Reset the list of all processors and initiate the walk over the main
+ provided processor package to load all available processors
+ """
+ self.processors = {}
+ self.seen_paths = []
+
+ logger.info(f'Loading processors from "{self.processors_dir}"...')
+ self.walk_package(self.processors_dir)
+
+ def walk_package(self, package):
+ """walk the supplied package to retrieve all processors"""
+ imported_package = __import__(package, fromlist=["blah"])
+ loaded_packages = []
+ for _, processor_name, ispkg in pkgutil.walk_packages(
+ imported_package.__path__, imported_package.__name__ + "."
+ ):
+ if not ispkg and processor_name != __name__:
+ processor_module = __import__(processor_name, fromlist=["blah"])
+ # https://stackoverflow.com/a/46206754/6242649
+ clsmembers = inspect.getmembers(
+ processor_module,
+ ProcessorManager.get_name_filter(processor_name),
+ )
+ for _, c in clsmembers:
+ # Only add classes that are a sub class of Processor, but NOT Processor itself
+ if issubclass(c, Processor) & (c is not Processor):
+ self.processors[c.__name__] = c
+ loaded_packages.append(c.__name__)
+
+ logger.info(f"Loaded processors: {loaded_packages}")
+
+
+# Singleton export
+PROCESSOR_MANAGER = ProcessorManager()
diff --git a/src/schemas/__init__.py b/src/schemas/__init__.py
new file mode 100644
index 00000000..3ccb298d
--- /dev/null
+++ b/src/schemas/__init__.py
@@ -0,0 +1,18 @@
+# https://docs.python.org/3/tutorial/modules.html#:~:text=The%20__init__.py,on%20the%20module%20search%20path.
+from jsonschema import Draft202012Validator
+
+from src.schemas.config_schema import CONFIG_SCHEMA
+from src.schemas.evaluation_schema import EVALUATION_SCHEMA
+from src.schemas.template_schema import TEMPLATE_SCHEMA
+
+SCHEMA_JSONS = {
+ "config": CONFIG_SCHEMA,
+ "evaluation": EVALUATION_SCHEMA,
+ "template": TEMPLATE_SCHEMA,
+}
+
+SCHEMA_VALIDATORS = {
+ "config": Draft202012Validator(CONFIG_SCHEMA),
+ "evaluation": Draft202012Validator(EVALUATION_SCHEMA),
+ "template": Draft202012Validator(TEMPLATE_SCHEMA),
+}
diff --git a/src/schemas/config_schema.py b/src/schemas/config_schema.py
new file mode 100644
index 00000000..6eb815f8
--- /dev/null
+++ b/src/schemas/config_schema.py
@@ -0,0 +1,57 @@
+CONFIG_SCHEMA = {
+ "$schema": "https://json-schema.org/draft/2020-12/schema",
+ "$id": "https://github.com/Udayraj123/OMRChecker/tree/master/src/schemas/config-schema.json",
+ "title": "Config Schema",
+ "description": "OMRChecker config schema for custom tuning",
+ "type": "object",
+ "additionalProperties": False,
+ "properties": {
+ "dimensions": {
+ "type": "object",
+ "additionalProperties": False,
+ "properties": {
+ "display_height": {"type": "integer"},
+ "display_width": {"type": "integer"},
+ "processing_height": {"type": "integer"},
+ "processing_width": {"type": "integer"},
+ },
+ },
+ "threshold_params": {
+ "type": "object",
+ "additionalProperties": False,
+ "properties": {
+ "GAMMA_LOW": {"type": "number", "minimum": 0, "maximum": 1},
+ "MIN_GAP": {"type": "integer", "minimum": 10, "maximum": 100},
+ "MIN_JUMP": {"type": "integer", "minimum": 10, "maximum": 100},
+ "CONFIDENT_SURPLUS": {"type": "integer", "minimum": 0, "maximum": 20},
+ "JUMP_DELTA": {"type": "integer", "minimum": 10, "maximum": 100},
+ "PAGE_TYPE_FOR_THRESHOLD": {
+ "enum": ["white", "black"],
+ "type": "string",
+ },
+ },
+ },
+ "alignment_params": {
+ "type": "object",
+ "additionalProperties": False,
+ "properties": {
+ "auto_align": {"type": "boolean"},
+ "match_col": {"type": "integer", "minimum": 0, "maximum": 10},
+ "max_steps": {"type": "integer", "minimum": 1, "maximum": 100},
+ "stride": {"type": "integer", "minimum": 1, "maximum": 10},
+ "thickness": {"type": "integer", "minimum": 1, "maximum": 10},
+ },
+ },
+ "outputs": {
+ "type": "object",
+ "additionalProperties": False,
+ "properties": {
+ "show_image_level": {"type": "integer", "minimum": 0, "maximum": 6},
+ "save_image_level": {"type": "integer", "minimum": 0, "maximum": 6},
+ "save_detections": {"type": "boolean"},
+ # This option moves multimarked files into a separate folder for manual checking, skipping evaluation
+ "filter_out_multimarked_files": {"type": "boolean"},
+ },
+ },
+ },
+}
diff --git a/src/schemas/constants.py b/src/schemas/constants.py
new file mode 100644
index 00000000..ccaa6f38
--- /dev/null
+++ b/src/schemas/constants.py
@@ -0,0 +1,17 @@
+DEFAULT_SECTION_KEY = "DEFAULT"
+
+BONUS_SECTION_PREFIX = "BONUS"
+
+MARKING_VERDICT_TYPES = ["correct", "incorrect", "unmarked"]
+
+ARRAY_OF_STRINGS = {
+ "type": "array",
+ "items": {"type": "string"},
+}
+
+FIELD_STRING_TYPE = {
+ "type": "string",
+ "pattern": "^([^\\.]+|[^\\.\\d]+\\d+\\.{2,3}\\d+)$",
+}
+
+FIELD_STRING_REGEX_GROUPS = r"([^\.\d]+)(\d+)\.{2,3}(\d+)"
diff --git a/src/schemas/evaluation_schema.py b/src/schemas/evaluation_schema.py
new file mode 100644
index 00000000..5f2237fd
--- /dev/null
+++ b/src/schemas/evaluation_schema.py
@@ -0,0 +1,151 @@
+from src.schemas.constants import (
+ ARRAY_OF_STRINGS,
+ DEFAULT_SECTION_KEY,
+ FIELD_STRING_TYPE,
+)
+
+marking_score_regex = "-?(\\d+)(/(\\d+))?"
+
+marking_score = {
+ "oneOf": [
+ {"type": "string", "pattern": marking_score_regex},
+ {"type": "number"},
+ ]
+}
+
+marking_object_properties = {
+ "additionalProperties": False,
+ "required": ["correct", "incorrect", "unmarked"],
+ "type": "object",
+ "properties": {
+ # TODO: can support streak marking if we allow array of marking_scores here
+ "correct": marking_score,
+ "incorrect": marking_score,
+ "unmarked": marking_score,
+ },
+}
+
+EVALUATION_SCHEMA = {
+ "$schema": "https://json-schema.org/draft/2020-12/schema",
+ "$id": "https://github.com/Udayraj123/OMRChecker/tree/master/src/schemas/evaluation-schema.json",
+ "title": "Evaluation Schema",
+ "description": "OMRChecker evaluation schema i.e. the marking scheme",
+ "type": "object",
+ "additionalProperties": True,
+ "required": ["source_type", "options", "marking_schemes"],
+ "properties": {
+ "additionalProperties": False,
+ "source_type": {"type": "string", "enum": ["csv", "custom"]},
+ "options": {"type": "object"},
+ "marking_schemes": {
+ "type": "object",
+ "required": [DEFAULT_SECTION_KEY],
+ "patternProperties": {
+ f"^{DEFAULT_SECTION_KEY}$": marking_object_properties,
+ f"^(?!{DEFAULT_SECTION_KEY}$).*": {
+ "additionalProperties": False,
+ "required": ["marking", "questions"],
+ "type": "object",
+ "properties": {
+ "questions": {
+ "oneOf": [
+ FIELD_STRING_TYPE,
+ {
+ "type": "array",
+ "items": FIELD_STRING_TYPE,
+ },
+ ]
+ },
+ "marking": marking_object_properties,
+ },
+ },
+ },
+ },
+ },
+ "allOf": [
+ {
+ "if": {"properties": {"source_type": {"const": "csv"}}},
+ "then": {
+ "properties": {
+ "options": {
+ "additionalProperties": False,
+ "required": ["answer_key_csv_path"],
+ "dependentRequired": {
+ "answer_key_image_path": [
+ "answer_key_csv_path",
+ "questions_in_order",
+ ]
+ },
+ "type": "object",
+ "properties": {
+ "should_explain_scoring": {"type": "boolean"},
+ "answer_key_csv_path": {"type": "string"},
+ "answer_key_image_path": {"type": "string"},
+ "questions_in_order": ARRAY_OF_STRINGS,
+ },
+ }
+ }
+ },
+ },
+ {
+ "if": {"properties": {"source_type": {"const": "custom"}}},
+ "then": {
+ "properties": {
+ "options": {
+ "additionalProperties": False,
+ "required": ["answers_in_order", "questions_in_order"],
+ "type": "object",
+ "properties": {
+ "should_explain_scoring": {"type": "boolean"},
+ "answers_in_order": {
+ "oneOf": [
+ {
+ "type": "array",
+ "items": {
+ "oneOf": [
+ # "standard": single correct, multi-marked single-correct
+ # Example: "q1" --> '67'
+ {"type": "string"},
+ # "multiple-correct": multiple-correct (for ambiguous/bonus questions)
+ # Example: "q1" --> [ 'A', 'B' ]
+ {
+ "type": "array",
+ "items": {"type": "string"},
+ "minItems": 2,
+ },
+ # "multiple-correct-weighted": array of answer-wise weights (marking scheme not applicable)
+ # Example 1: "q1" --> [['A', 1], ['B', 2], ['C', 3]] or
+ # Example 2: "q2" --> [['A', 1], ['B', 1], ['AB', 2]]
+ {
+ "type": "array",
+ "items": {
+ "type": "array",
+ "items": False,
+ "minItems": 2,
+ "maxItems": 2,
+ "prefixItems": [
+ {"type": "string"},
+ marking_score,
+ ],
+ },
+ },
+ # Multiple-correct with custom marking scheme
+ # ["A", ["1", "2", "3"]],
+ # [["A", "B", "AB"], ["1", "2", "3"]]
+ ],
+ },
+ },
+ ]
+ },
+ "questions_in_order": ARRAY_OF_STRINGS,
+ "enable_evaluation_table_to_csv": {
+ "type": "boolean",
+ "default": False,
+ },
+ },
+ }
+ }
+ },
+ },
+ ],
+}
diff --git a/src/schemas/template_schema.py b/src/schemas/template_schema.py
new file mode 100644
index 00000000..451d499d
--- /dev/null
+++ b/src/schemas/template_schema.py
@@ -0,0 +1,226 @@
+from src.constants import FIELD_TYPES
+from src.schemas.constants import ARRAY_OF_STRINGS, FIELD_STRING_TYPE
+
+positive_number = {"type": "number", "minimum": 0}
+positive_integer = {"type": "integer", "minimum": 0}
+two_positive_integers = {
+ "type": "array",
+ "prefixItems": [
+ positive_integer,
+ positive_integer,
+ ],
+ "maxItems": 2,
+ "minItems": 2,
+}
+two_positive_numbers = {
+ "type": "array",
+ "prefixItems": [
+ positive_number,
+ positive_number,
+ ],
+ "maxItems": 2,
+ "minItems": 2,
+}
+zero_to_one_number = {
+ "type": "number",
+ "minimum": 0,
+ "maximum": 1,
+}
+
+TEMPLATE_SCHEMA = {
+ "$schema": "https://json-schema.org/draft/2020-12/schema",
+ "$id": "https://github.com/Udayraj123/OMRChecker/tree/master/src/schemas/template-schema.json",
+ "title": "Template Validation Schema",
+ "description": "OMRChecker input template schema",
+ "type": "object",
+ "required": [
+ "bubbleDimensions",
+ "pageDimensions",
+ "preProcessors",
+ "fieldBlocks",
+ ],
+ "additionalProperties": False,
+ "properties": {
+ "bubbleDimensions": {
+ **two_positive_integers,
+ "description": "The dimensions of the overlay bubble area: [width, height]",
+ },
+ "customLabels": {
+ "description": "The customLabels contain fields that need to be joined together before generating the results sheet",
+ "type": "object",
+ "patternProperties": {
+ "^.*$": {"type": "array", "items": FIELD_STRING_TYPE}
+ },
+ },
+ "outputColumns": {
+ "type": "array",
+ "items": FIELD_STRING_TYPE,
+ "description": "The ordered list of columns to be contained in the output csv(default order: alphabetical)",
+ },
+ "pageDimensions": {
+ **two_positive_integers,
+ "description": "The dimensions(width, height) to which the page will be resized to before applying template",
+ },
+ "preProcessors": {
+ "description": "Custom configuration values to use in the template's directory",
+ "type": "array",
+ "items": {
+ "type": "object",
+ "properties": {
+ "name": {
+ "type": "string",
+ "enum": [
+ "CropOnMarkers",
+ "CropPage",
+ "FeatureBasedAlignment",
+ "GaussianBlur",
+ "Levels",
+ "MedianBlur",
+ ],
+ },
+ },
+ "required": ["name", "options"],
+ "allOf": [
+ {
+ "if": {"properties": {"name": {"const": "CropOnMarkers"}}},
+ "then": {
+ "properties": {
+ "options": {
+ "type": "object",
+ "additionalProperties": False,
+ "properties": {
+ "apply_erode_subtract": {"type": "boolean"},
+ "marker_rescale_range": two_positive_numbers,
+ "marker_rescale_steps": {"type": "number"},
+ "max_matching_variation": {"type": "number"},
+ "min_matching_threshold": {"type": "number"},
+ "relativePath": {"type": "string"},
+ "sheetToMarkerWidthRatio": {"type": "number"},
+ },
+ "required": ["relativePath"],
+ }
+ }
+ },
+ },
+ {
+ "if": {
+ "properties": {"name": {"const": "FeatureBasedAlignment"}}
+ },
+ "then": {
+ "properties": {
+ "options": {
+ "type": "object",
+ "additionalProperties": False,
+ "properties": {
+ "2d": {"type": "boolean"},
+ "goodMatchPercent": {"type": "number"},
+ "maxFeatures": {"type": "integer"},
+ "reference": {"type": "string"},
+ },
+ "required": ["reference"],
+ }
+ }
+ },
+ },
+ {
+ "if": {"properties": {"name": {"const": "Levels"}}},
+ "then": {
+ "properties": {
+ "options": {
+ "type": "object",
+ "additionalProperties": False,
+ "properties": {
+ "gamma": zero_to_one_number,
+ "high": zero_to_one_number,
+ "low": zero_to_one_number,
+ },
+ }
+ }
+ },
+ },
+ {
+ "if": {"properties": {"name": {"const": "MedianBlur"}}},
+ "then": {
+ "properties": {
+ "options": {
+ "type": "object",
+ "additionalProperties": False,
+ "properties": {"kSize": {"type": "integer"}},
+ }
+ }
+ },
+ },
+ {
+ "if": {"properties": {"name": {"const": "GaussianBlur"}}},
+ "then": {
+ "properties": {
+ "options": {
+ "type": "object",
+ "additionalProperties": False,
+ "properties": {
+ "kSize": two_positive_integers,
+ "sigmaX": {"type": "number"},
+ },
+ }
+ }
+ },
+ },
+ {
+ "if": {"properties": {"name": {"const": "CropPage"}}},
+ "then": {
+ "properties": {
+ "options": {
+ "type": "object",
+ "additionalProperties": False,
+ "properties": {
+ "morphKernel": two_positive_integers
+ },
+ }
+ }
+ },
+ },
+ ],
+ },
+ },
+ "fieldBlocks": {
+ "description": "The fieldBlocks denote small groups of adjacent fields",
+ "type": "object",
+ "patternProperties": {
+ "^.*$": {
+ "type": "object",
+ "required": [
+ "origin",
+ "bubblesGap",
+ "labelsGap",
+ "fieldLabels",
+ ],
+ "oneOf": [
+ {"required": ["fieldType"]},
+ {"required": ["bubbleValues", "direction"]},
+ ],
+ "properties": {
+ "bubbleDimensions": two_positive_numbers,
+ "bubblesGap": positive_number,
+ "bubbleValues": ARRAY_OF_STRINGS,
+ "direction": {
+ "type": "string",
+ "enum": ["horizontal", "vertical"],
+ },
+ "emptyValue": {"type": "string"},
+ "fieldLabels": {"type": "array", "items": FIELD_STRING_TYPE},
+ "labelsGap": positive_number,
+ "origin": two_positive_integers,
+ "fieldType": {
+ "type": "string",
+ "enum": list(FIELD_TYPES.keys()),
+ },
+ },
+ }
+ },
+ },
+ "emptyValue": {
+ "description": "The value to be used in case of empty bubble detected at global level.",
+ "type": "string",
+ },
+ },
+}
diff --git a/src/template.py b/src/template.py
new file mode 100644
index 00000000..0acc4672
--- /dev/null
+++ b/src/template.py
@@ -0,0 +1,327 @@
+"""
+
+ OMRChecker
+
+ Author: Udayraj Deshmukh
+ Github: https://github.com/Udayraj123
+
+"""
+from src.constants import FIELD_TYPES
+from src.core import ImageInstanceOps
+from src.logger import logger
+from src.processors.manager import PROCESSOR_MANAGER
+from src.utils.parsing import (
+ custom_sort_output_columns,
+ open_template_with_defaults,
+ parse_fields,
+)
+
+
+class Template:
+ def __init__(self, template_path, tuning_config):
+ self.path = template_path
+ self.image_instance_ops = ImageInstanceOps(tuning_config)
+
+ json_object = open_template_with_defaults(template_path)
+ (
+ custom_labels_object,
+ field_blocks_object,
+ output_columns_array,
+ pre_processors_object,
+ self.bubble_dimensions,
+ self.global_empty_val,
+ self.options,
+ self.page_dimensions,
+ ) = map(
+ json_object.get,
+ [
+ "customLabels",
+ "fieldBlocks",
+ "outputColumns",
+ "preProcessors",
+ "bubbleDimensions",
+ "emptyValue",
+ "options",
+ "pageDimensions",
+ ],
+ )
+
+ self.parse_output_columns(output_columns_array)
+ self.setup_pre_processors(pre_processors_object, template_path.parent)
+ self.setup_field_blocks(field_blocks_object)
+ self.parse_custom_labels(custom_labels_object)
+
+ non_custom_columns, all_custom_columns = (
+ list(self.non_custom_labels),
+ list(custom_labels_object.keys()),
+ )
+
+ if len(self.output_columns) == 0:
+ self.fill_output_columns(non_custom_columns, all_custom_columns)
+
+ self.validate_template_columns(non_custom_columns, all_custom_columns)
+
+ def parse_output_columns(self, output_columns_array):
+ self.output_columns = parse_fields(f"Output Columns", output_columns_array)
+
+ def setup_pre_processors(self, pre_processors_object, relative_dir):
+ # load image pre_processors
+ self.pre_processors = []
+ for pre_processor in pre_processors_object:
+ ProcessorClass = PROCESSOR_MANAGER.processors[pre_processor["name"]]
+ pre_processor_instance = ProcessorClass(
+ options=pre_processor["options"],
+ relative_dir=relative_dir,
+ image_instance_ops=self.image_instance_ops,
+ )
+ self.pre_processors.append(pre_processor_instance)
+
+ def setup_field_blocks(self, field_blocks_object):
+ # Add field_blocks
+ self.field_blocks = []
+ self.all_parsed_labels = set()
+ for block_name, field_block_object in field_blocks_object.items():
+ self.parse_and_add_field_block(block_name, field_block_object)
+
+ def parse_custom_labels(self, custom_labels_object):
+ all_parsed_custom_labels = set()
+ self.custom_labels = {}
+ for custom_label, label_strings in custom_labels_object.items():
+ parsed_labels = parse_fields(f"Custom Label: {custom_label}", label_strings)
+ parsed_labels_set = set(parsed_labels)
+ self.custom_labels[custom_label] = parsed_labels
+
+ missing_custom_labels = sorted(
+ parsed_labels_set.difference(self.all_parsed_labels)
+ )
+ if len(missing_custom_labels) > 0:
+ logger.critical(
+ f"For '{custom_label}', Missing labels - {missing_custom_labels}"
+ )
+ raise Exception(
+ f"Missing field block label(s) in the given template for {missing_custom_labels} from '{custom_label}'"
+ )
+
+ if not all_parsed_custom_labels.isdisjoint(parsed_labels_set):
+ # Note: this can be made a warning, but it's a choice
+ logger.critical(
+ f"field strings overlap for labels: {label_strings} and existing custom labels: {all_parsed_custom_labels}"
+ )
+ raise Exception(
+ f"The field strings for custom label '{custom_label}' overlap with other existing custom labels"
+ )
+
+ all_parsed_custom_labels.update(parsed_labels)
+
+ self.non_custom_labels = self.all_parsed_labels.difference(
+ all_parsed_custom_labels
+ )
+
+ def fill_output_columns(self, non_custom_columns, all_custom_columns):
+ all_template_columns = non_custom_columns + all_custom_columns
+ # Typical case: sort alpha-numerical (natural sort)
+ self.output_columns = sorted(
+ all_template_columns, key=custom_sort_output_columns
+ )
+
+ def validate_template_columns(self, non_custom_columns, all_custom_columns):
+ output_columns_set = set(self.output_columns)
+ all_custom_columns_set = set(all_custom_columns)
+
+ missing_output_columns = sorted(
+ output_columns_set.difference(all_custom_columns_set).difference(
+ self.all_parsed_labels
+ )
+ )
+ if len(missing_output_columns) > 0:
+ logger.critical(f"Missing output columns: {missing_output_columns}")
+ raise Exception(
+ f"Some columns are missing in the field blocks for the given output columns"
+ )
+
+ all_template_columns_set = set(non_custom_columns + all_custom_columns)
+ missing_label_columns = sorted(
+ all_template_columns_set.difference(output_columns_set)
+ )
+ if len(missing_label_columns) > 0:
+ logger.warning(
+ f"Some label columns are not covered in the given output columns: {missing_label_columns}"
+ )
+
+ def parse_and_add_field_block(self, block_name, field_block_object):
+ field_block_object = self.pre_fill_field_block(field_block_object)
+ block_instance = FieldBlock(block_name, field_block_object)
+ self.field_blocks.append(block_instance)
+ self.validate_parsed_labels(field_block_object["fieldLabels"], block_instance)
+
+ def pre_fill_field_block(self, field_block_object):
+ if "fieldType" in field_block_object:
+ field_block_object = {
+ **field_block_object,
+ **FIELD_TYPES[field_block_object["fieldType"]],
+ }
+ else:
+ field_block_object = {**field_block_object, "fieldType": "__CUSTOM__"}
+
+ return {
+ "direction": "vertical",
+ "emptyValue": self.global_empty_val,
+ "bubbleDimensions": self.bubble_dimensions,
+ **field_block_object,
+ }
+
+ def validate_parsed_labels(self, field_labels, block_instance):
+ parsed_field_labels, block_name = (
+ block_instance.parsed_field_labels,
+ block_instance.name,
+ )
+ field_labels_set = set(parsed_field_labels)
+ if not self.all_parsed_labels.isdisjoint(field_labels_set):
+ # Note: in case of two fields pointing to same column, use a custom column instead of same field labels.
+ logger.critical(
+ f"An overlap found between field string: {field_labels} in block '{block_name}' and existing labels: {self.all_parsed_labels}"
+ )
+ raise Exception(
+ f"The field strings for field block {block_name} overlap with other existing fields"
+ )
+ self.all_parsed_labels.update(field_labels_set)
+
+ page_width, page_height = self.page_dimensions
+ block_width, block_height = block_instance.dimensions
+ [block_start_x, block_start_y] = block_instance.origin
+
+ block_end_x, block_end_y = (
+ block_start_x + block_width,
+ block_start_y + block_height,
+ )
+
+ if (
+ block_end_x >= page_width
+ or block_end_y >= page_height
+ or block_start_x < 0
+ or block_start_y < 0
+ ):
+ raise Exception(
+ f"Overflowing field block '{block_name}' with origin {block_instance.origin} and dimensions {block_instance.dimensions} in template with dimensions {self.page_dimensions}"
+ )
+
+ def __str__(self):
+ return str(self.path)
+
+
+class FieldBlock:
+ def __init__(self, block_name, field_block_object):
+ self.name = block_name
+ self.shift = 0
+ self.setup_field_block(field_block_object)
+
+ def setup_field_block(self, field_block_object):
+ # case mapping
+ (
+ bubble_dimensions,
+ bubble_values,
+ bubbles_gap,
+ direction,
+ field_labels,
+ field_type,
+ labels_gap,
+ origin,
+ self.empty_val,
+ ) = map(
+ field_block_object.get,
+ [
+ "bubbleDimensions",
+ "bubbleValues",
+ "bubblesGap",
+ "direction",
+ "fieldLabels",
+ "fieldType",
+ "labelsGap",
+ "origin",
+ "emptyValue",
+ ],
+ )
+ self.parsed_field_labels = parse_fields(
+ f"Field Block Labels: {self.name}", field_labels
+ )
+ self.origin = origin
+ self.bubble_dimensions = bubble_dimensions
+ self.calculate_block_dimensions(
+ bubble_dimensions,
+ bubble_values,
+ bubbles_gap,
+ direction,
+ labels_gap,
+ )
+ self.generate_bubble_grid(
+ bubble_values,
+ bubbles_gap,
+ direction,
+ field_type,
+ labels_gap,
+ )
+
+ def calculate_block_dimensions(
+ self,
+ bubble_dimensions,
+ bubble_values,
+ bubbles_gap,
+ direction,
+ labels_gap,
+ ):
+ _h, _v = (1, 0) if (direction == "vertical") else (0, 1)
+
+ values_dimension = int(
+ bubbles_gap * (len(bubble_values) - 1) + bubble_dimensions[_h]
+ )
+ fields_dimension = int(
+ labels_gap * (len(self.parsed_field_labels) - 1) + bubble_dimensions[_v]
+ )
+ self.dimensions = (
+ [fields_dimension, values_dimension]
+ if (direction == "vertical")
+ else [values_dimension, fields_dimension]
+ )
+
+ def generate_bubble_grid(
+ self,
+ bubble_values,
+ bubbles_gap,
+ direction,
+ field_type,
+ labels_gap,
+ ):
+ _h, _v = (1, 0) if (direction == "vertical") else (0, 1)
+ self.traverse_bubbles = []
+ # Generate the bubble grid
+ lead_point = [float(self.origin[0]), float(self.origin[1])]
+ for field_label in self.parsed_field_labels:
+ bubble_point = lead_point.copy()
+ field_bubbles = []
+ for bubble_value in bubble_values:
+ field_bubbles.append(
+ Bubble(bubble_point.copy(), field_label, field_type, bubble_value)
+ )
+ bubble_point[_h] += bubbles_gap
+ self.traverse_bubbles.append(field_bubbles)
+ lead_point[_v] += labels_gap
+
+
+class Bubble:
+ """
+ Container for a Point Box on the OMR
+
+ field_label is the point's property- field to which this point belongs to
+ It can be used as a roll number column as well. (eg roll1)
+ It can also correspond to a single digit of integer type Q (eg q5d1)
+ """
+
+ def __init__(self, pt, field_label, field_type, field_value):
+ self.x = round(pt[0])
+ self.y = round(pt[1])
+ self.field_label = field_label
+ self.field_type = field_type
+ self.field_value = field_value
+
+ def __str__(self):
+ return str([self.x, self.y])
diff --git a/src/tests/__init__.py b/src/tests/__init__.py
new file mode 100644
index 00000000..855c16a5
--- /dev/null
+++ b/src/tests/__init__.py
@@ -0,0 +1 @@
+# https://stackoverflow.com/a/50169991/6242649
diff --git a/src/tests/__snapshots__/test_all_samples.ambr b/src/tests/__snapshots__/test_all_samples.ambr
new file mode 100644
index 00000000..3049cd3c
--- /dev/null
+++ b/src/tests/__snapshots__/test_all_samples.ambr
@@ -0,0 +1,302 @@
+# serializer version: 1
+# name: test_run_answer_key_using_csv
+ dict({
+ 'Manual/ErrorFiles.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5"
+
+ ''',
+ 'Manual/MultiMarkedFiles.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5"
+
+ ''',
+ 'Results/Results_05AM.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5"
+ "adrian_omr.png","samples/answer-key/using-csv/adrian_omr.png","outputs/answer-key/using-csv/CheckedOMRs/adrian_omr.png","5.0","C","E","A","B","B"
+
+ ''',
+ })
+# ---
+# name: test_run_answer_key_weighted_answers
+ dict({
+ 'images/Manual/ErrorFiles.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5"
+
+ ''',
+ 'images/Manual/MultiMarkedFiles.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5"
+
+ ''',
+ 'images/Results/Results_05AM.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5"
+ "adrian_omr.png","samples/answer-key/weighted-answers/images/adrian_omr.png","outputs/answer-key/weighted-answers/images/CheckedOMRs/adrian_omr.png","5.5","B","E","A","C","B"
+ "adrian_omr_2.png","samples/answer-key/weighted-answers/images/adrian_omr_2.png","outputs/answer-key/weighted-answers/images/CheckedOMRs/adrian_omr_2.png","10.0","C","E","A","B","B"
+
+ ''',
+ })
+# ---
+# name: test_run_community_Antibodyy
+ dict({
+ 'Manual/ErrorFiles.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5","q6"
+
+ ''',
+ 'Manual/MultiMarkedFiles.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5","q6"
+
+ ''',
+ 'Results/Results_05AM.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5","q6"
+ "simple_omr_sheet.jpg","samples/community/Antibodyy/simple_omr_sheet.jpg","outputs/community/Antibodyy/CheckedOMRs/simple_omr_sheet.jpg","0","A","C","B","D","E","B"
+
+ ''',
+ })
+# ---
+# name: test_run_community_Sandeep_1507
+ dict({
+ 'Manual/ErrorFiles.csv': '''
+ "file_id","input_path","output_path","score","Booklet_No","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22","q23","q24","q25","q26","q27","q28","q29","q30","q31","q32","q33","q34","q35","q36","q37","q38","q39","q40","q41","q42","q43","q44","q45","q46","q47","q48","q49","q50","q51","q52","q53","q54","q55","q56","q57","q58","q59","q60","q61","q62","q63","q64","q65","q66","q67","q68","q69","q70","q71","q72","q73","q74","q75","q76","q77","q78","q79","q80","q81","q82","q83","q84","q85","q86","q87","q88","q89","q90","q91","q92","q93","q94","q95","q96","q97","q98","q99","q100","q101","q102","q103","q104","q105","q106","q107","q108","q109","q110","q111","q112","q113","q114","q115","q116","q117","q118","q119","q120","q121","q122","q123","q124","q125","q126","q127","q128","q129","q130","q131","q132","q133","q134","q135","q136","q137","q138","q139","q140","q141","q142","q143","q144","q145","q146","q147","q148","q149","q150","q151","q152","q153","q154","q155","q156","q157","q158","q159","q160","q161","q162","q163","q164","q165","q166","q167","q168","q169","q170","q171","q172","q173","q174","q175","q176","q177","q178","q179","q180","q181","q182","q183","q184","q185","q186","q187","q188","q189","q190","q191","q192","q193","q194","q195","q196","q197","q198","q199","q200"
+
+ ''',
+ 'Manual/MultiMarkedFiles.csv': '''
+ "file_id","input_path","output_path","score","Booklet_No","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22","q23","q24","q25","q26","q27","q28","q29","q30","q31","q32","q33","q34","q35","q36","q37","q38","q39","q40","q41","q42","q43","q44","q45","q46","q47","q48","q49","q50","q51","q52","q53","q54","q55","q56","q57","q58","q59","q60","q61","q62","q63","q64","q65","q66","q67","q68","q69","q70","q71","q72","q73","q74","q75","q76","q77","q78","q79","q80","q81","q82","q83","q84","q85","q86","q87","q88","q89","q90","q91","q92","q93","q94","q95","q96","q97","q98","q99","q100","q101","q102","q103","q104","q105","q106","q107","q108","q109","q110","q111","q112","q113","q114","q115","q116","q117","q118","q119","q120","q121","q122","q123","q124","q125","q126","q127","q128","q129","q130","q131","q132","q133","q134","q135","q136","q137","q138","q139","q140","q141","q142","q143","q144","q145","q146","q147","q148","q149","q150","q151","q152","q153","q154","q155","q156","q157","q158","q159","q160","q161","q162","q163","q164","q165","q166","q167","q168","q169","q170","q171","q172","q173","q174","q175","q176","q177","q178","q179","q180","q181","q182","q183","q184","q185","q186","q187","q188","q189","q190","q191","q192","q193","q194","q195","q196","q197","q198","q199","q200"
+
+ ''',
+ 'Results/Results_05AM.csv': '''
+ "file_id","input_path","output_path","score","Booklet_No","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22","q23","q24","q25","q26","q27","q28","q29","q30","q31","q32","q33","q34","q35","q36","q37","q38","q39","q40","q41","q42","q43","q44","q45","q46","q47","q48","q49","q50","q51","q52","q53","q54","q55","q56","q57","q58","q59","q60","q61","q62","q63","q64","q65","q66","q67","q68","q69","q70","q71","q72","q73","q74","q75","q76","q77","q78","q79","q80","q81","q82","q83","q84","q85","q86","q87","q88","q89","q90","q91","q92","q93","q94","q95","q96","q97","q98","q99","q100","q101","q102","q103","q104","q105","q106","q107","q108","q109","q110","q111","q112","q113","q114","q115","q116","q117","q118","q119","q120","q121","q122","q123","q124","q125","q126","q127","q128","q129","q130","q131","q132","q133","q134","q135","q136","q137","q138","q139","q140","q141","q142","q143","q144","q145","q146","q147","q148","q149","q150","q151","q152","q153","q154","q155","q156","q157","q158","q159","q160","q161","q162","q163","q164","q165","q166","q167","q168","q169","q170","q171","q172","q173","q174","q175","q176","q177","q178","q179","q180","q181","q182","q183","q184","q185","q186","q187","q188","q189","q190","q191","q192","q193","q194","q195","q196","q197","q198","q199","q200"
+ "omr-1.png","samples/community/Sandeep-1507/omr-1.png","outputs/community/Sandeep-1507/CheckedOMRs/omr-1.png","0","0190880","D","C","B","A","A","B","C","D","D","C","B","A","D","A","B","C","D","","B","D","C","A","C","C","B","A","D","A","AC","C","B","D","C","B","A","B","B","D","D","A","C","B","D","A","C","B","D","B","D","A","A","B","C","D","C","B","A","D","D","A","B","C","D","C","B","A","B","C","D","A","B","C","D","B","A","C","D","C","B","A","D","B","D","A","A","B","A","C","B","D","C","D","B","A","C","C","B","D","B","C","B","A","D","C","B","A","B","C","D","A","A","A","B","B","A","B","C","D","A","A","D","C","B","A","","A","B","C","D","D","D","B","B","C","C","D","C","C","D","D","C","C","B","B","A","A","D","D","B","A","D","C","B","A","A","D","D","B","B","A","A","B","C","D","D","C","B","A","B","D","A","C","C","C","A","A","B","B","D","D","A","A","B","C","D","B","D","A","B","C","D","AD","C","D","B","C","A","B","C","D"
+ "omr-2.png","samples/community/Sandeep-1507/omr-2.png","outputs/community/Sandeep-1507/CheckedOMRs/omr-2.png","0","0no22nonono","A","B","B","A","D","C","B","D","C","D","D","D","B","B","D","D","D","B","C","C","A","A","B","A","D","A","A","B","A","C","A","C","D","D","D","","","C","C","B","B","B","","D","","C","D","","D","B","A","D","B","A","C","A","C","A","C","B","A","D","C","B","C","B","C","D","B","B","D","C","C","D","D","A","D","A","D","C","B","D","C","A","C","","C","B","B","","A","A","D","","B","A","","C","A","D","D","C","C","A","C","A","C","D","A","A","A","D","D","B","C","B","B","B","D","A","C","D","D","A","A","A","C","D","C","C","B","D","A","A","C","B","","D","A","C","C","C","","","","A","C","","D","A","B","A","A","C","A","D","B","B","A","D","A","B","C","A","C","D","D","D","C","A","C","A","C","D","A","A","A","D","A","B","A","B","C","B","A","","B","C","D","D","","","D","C","C","C","","C","A",""
+ "omr-3.png","samples/community/Sandeep-1507/omr-3.png","outputs/community/Sandeep-1507/CheckedOMRs/omr-3.png","0","0nononono73","B","A","C","D","A","D","D","A","C","A","A","B","C","A","A","C","A","B","A","D","C","C","A","D","D","C","C","C","A","C","C","B","B","D","D","C","","","C","B","","","D","A","A","A","A","","A","C","C","C","D","C","","A","B","C","D","B","C","C","C","D","A","B","B","B","D","D","B","B","C","D","B","D","A","B","A","B","C","A","C","A","C","D","","","A","B","","B","C","D","A","D","D","","","C","D","B","B","A","A","D","D","B","A","B","B","C","C","D","D","C","A","D","C","D","C","C","B","C","D","C","D","A","B","D","C","B","D","B","B","","D","","B","D","B","B","C","A","D","","C","","C","","B","C","A","B","B","D","D","D","B","A","D","D","A","D","D","C","B","B","D","C","B","A","C","D","A","D","D","A","C","A","B","D","C","C","C","A","D","","","B","B","","C","C","B","B","C","","","B"
+
+ ''',
+ })
+# ---
+# name: test_run_community_Shamanth
+ dict({
+ 'Manual/ErrorFiles.csv': '''
+ "file_id","input_path","output_path","score","q21","q22","q23","q24","q25","q26","q27","q28"
+
+ ''',
+ 'Manual/MultiMarkedFiles.csv': '''
+ "file_id","input_path","output_path","score","q21","q22","q23","q24","q25","q26","q27","q28"
+
+ ''',
+ 'Results/Results_05AM.csv': '''
+ "file_id","input_path","output_path","score","q21","q22","q23","q24","q25","q26","q27","q28"
+ "omr_sheet_01.png","samples/community/Shamanth/omr_sheet_01.png","outputs/community/Shamanth/CheckedOMRs/omr_sheet_01.png","0","A","B","C","D","A","C","C","D"
+
+ ''',
+ })
+# ---
+# name: test_run_community_UPSC_mock
+ dict({
+ 'Manual/ErrorFiles.csv': '''
+ "file_id","input_path","output_path","score","Roll","Subject Code","bookletNo","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22","q23","q24","q25","q26","q27","q28","q29","q30","q31","q32","q33","q34","q35","q36","q37","q38","q39","q40","q41","q42","q43","q44","q45","q46","q47","q48","q49","q50","q51","q52","q53","q54","q55","q56","q57","q58","q59","q60","q61","q62","q63","q64","q65","q66","q67","q68","q69","q70","q71","q72","q73","q74","q75","q76","q77","q78","q79","q80","q81","q82","q83","q84","q85","q86","q87","q88","q89","q90","q91","q92","q93","q94","q95","q96","q97","q98","q99","q100"
+
+ ''',
+ 'Manual/MultiMarkedFiles.csv': '''
+ "file_id","input_path","output_path","score","Roll","Subject Code","bookletNo","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22","q23","q24","q25","q26","q27","q28","q29","q30","q31","q32","q33","q34","q35","q36","q37","q38","q39","q40","q41","q42","q43","q44","q45","q46","q47","q48","q49","q50","q51","q52","q53","q54","q55","q56","q57","q58","q59","q60","q61","q62","q63","q64","q65","q66","q67","q68","q69","q70","q71","q72","q73","q74","q75","q76","q77","q78","q79","q80","q81","q82","q83","q84","q85","q86","q87","q88","q89","q90","q91","q92","q93","q94","q95","q96","q97","q98","q99","q100"
+
+ ''',
+ 'Results/Results_05AM.csv': '''
+ "file_id","input_path","output_path","score","Roll","Subject Code","bookletNo","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22","q23","q24","q25","q26","q27","q28","q29","q30","q31","q32","q33","q34","q35","q36","q37","q38","q39","q40","q41","q42","q43","q44","q45","q46","q47","q48","q49","q50","q51","q52","q53","q54","q55","q56","q57","q58","q59","q60","q61","q62","q63","q64","q65","q66","q67","q68","q69","q70","q71","q72","q73","q74","q75","q76","q77","q78","q79","q80","q81","q82","q83","q84","q85","q86","q87","q88","q89","q90","q91","q92","q93","q94","q95","q96","q97","q98","q99","q100"
+ "answer_key.jpg","samples/community/UPSC-mock/answer_key.jpg","outputs/community/UPSC-mock/CheckedOMRs/answer_key.jpg","200.0","","","","C","D","A","C","C","C","B","A","C","C","B","D","B","D","C","C","B","D","B","D","C","C","C","B","D","D","D","B","A","D","D","C","A","B","C","A","D","A","A","A","D","D","B","A","B","C","B","A","C","D","C","D","A","B","C","A","C","C","C","D","B","C","C","C","C","A","D","A","D","A","D","C","C","D","C","D","A","A","C","B","C","D","C","A","B","C","B","D","A","A","C","A","B","D","C","D","A","C","B","A"
+
+ ''',
+ 'scan-angles/Manual/ErrorFiles.csv': '''
+ "file_id","input_path","output_path","score","Roll","Subject Code","bookletNo","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22","q23","q24","q25","q26","q27","q28","q29","q30","q31","q32","q33","q34","q35","q36","q37","q38","q39","q40","q41","q42","q43","q44","q45","q46","q47","q48","q49","q50","q51","q52","q53","q54","q55","q56","q57","q58","q59","q60","q61","q62","q63","q64","q65","q66","q67","q68","q69","q70","q71","q72","q73","q74","q75","q76","q77","q78","q79","q80","q81","q82","q83","q84","q85","q86","q87","q88","q89","q90","q91","q92","q93","q94","q95","q96","q97","q98","q99","q100"
+
+ ''',
+ 'scan-angles/Manual/MultiMarkedFiles.csv': '''
+ "file_id","input_path","output_path","score","Roll","Subject Code","bookletNo","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22","q23","q24","q25","q26","q27","q28","q29","q30","q31","q32","q33","q34","q35","q36","q37","q38","q39","q40","q41","q42","q43","q44","q45","q46","q47","q48","q49","q50","q51","q52","q53","q54","q55","q56","q57","q58","q59","q60","q61","q62","q63","q64","q65","q66","q67","q68","q69","q70","q71","q72","q73","q74","q75","q76","q77","q78","q79","q80","q81","q82","q83","q84","q85","q86","q87","q88","q89","q90","q91","q92","q93","q94","q95","q96","q97","q98","q99","q100"
+
+ ''',
+ 'scan-angles/Results/Results_05AM.csv': '''
+ "file_id","input_path","output_path","score","Roll","Subject Code","bookletNo","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22","q23","q24","q25","q26","q27","q28","q29","q30","q31","q32","q33","q34","q35","q36","q37","q38","q39","q40","q41","q42","q43","q44","q45","q46","q47","q48","q49","q50","q51","q52","q53","q54","q55","q56","q57","q58","q59","q60","q61","q62","q63","q64","q65","q66","q67","q68","q69","q70","q71","q72","q73","q74","q75","q76","q77","q78","q79","q80","q81","q82","q83","q84","q85","q86","q87","q88","q89","q90","q91","q92","q93","q94","q95","q96","q97","q98","q99","q100"
+ "angle-1.jpg","samples/community/UPSC-mock/scan-angles/angle-1.jpg","outputs/community/UPSC-mock/scan-angles/CheckedOMRs/angle-1.jpg","70.66666666666669","","","","D","D","A","","C","C","B","","A","C","C","D","A","D","A","C","A","D","B","D","D","C","D","D","D","D","","B","A","D","D","C","","B","","C","D","","","A","","A","C","C","B","C","A","A","C","","C","","D","B","C","","B","C","D","","","C","C","","C","A","B","C","","","","","D","D","C","D","A","","","B","","B","D","C","C","","D","","D","C","D","A","","A","","","A","C","B","A"
+ "angle-2.jpg","samples/community/UPSC-mock/scan-angles/angle-2.jpg","outputs/community/UPSC-mock/scan-angles/CheckedOMRs/angle-2.jpg","70.66666666666669","","","","D","D","A","","C","C","B","","A","C","C","D","A","D","A","C","A","D","B","D","D","C","D","D","D","D","","B","A","D","D","C","","B","","C","D","","","A","","A","C","C","B","C","A","A","C","","C","","D","B","C","","B","C","D","","","C","C","","C","A","B","C","","","","","D","D","C","D","A","","","B","","B","D","C","C","","D","","D","C","D","A","","A","","","A","C","B","A"
+ "angle-3.jpg","samples/community/UPSC-mock/scan-angles/angle-3.jpg","outputs/community/UPSC-mock/scan-angles/CheckedOMRs/angle-3.jpg","70.66666666666669","","","","D","D","A","","C","C","B","","A","C","C","D","A","D","A","C","A","D","B","D","D","C","D","D","D","D","","B","A","D","D","C","","B","","C","D","","","A","","A","C","C","B","C","A","A","C","","C","","D","B","C","","B","C","D","","","C","C","","C","A","B","C","","","","","D","D","C","D","A","","","B","","B","D","C","C","","D","","D","C","D","A","","A","","","A","C","B","A"
+
+ ''',
+ })
+# ---
+# name: test_run_community_UmarFarootAPS
+ dict({
+ 'scans/Manual/ErrorFiles.csv': '''
+ "file_id","input_path","output_path","score","Roll_no","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22","q23","q24","q25","q26","q27","q28","q29","q30","q31","q32","q33","q34","q35","q36","q37","q38","q39","q40","q41","q42","q43","q44","q45","q46","q47","q48","q49","q50","q51","q52","q53","q54","q55","q56","q57","q58","q59","q60","q61","q62","q63","q64","q65","q66","q67","q68","q69","q70","q71","q72","q73","q74","q75","q76","q77","q78","q79","q80","q81","q82","q83","q84","q85","q86","q87","q88","q89","q90","q91","q92","q93","q94","q95","q96","q97","q98","q99","q100","q101","q102","q103","q104","q105","q106","q107","q108","q109","q110","q111","q112","q113","q114","q115","q116","q117","q118","q119","q120","q121","q122","q123","q124","q125","q126","q127","q128","q129","q130","q131","q132","q133","q134","q135","q136","q137","q138","q139","q140","q141","q142","q143","q144","q145","q146","q147","q148","q149","q150","q151","q152","q153","q154","q155","q156","q157","q158","q159","q160","q161","q162","q163","q164","q165","q166","q167","q168","q169","q170","q171","q172","q173","q174","q175","q176","q177","q178","q179","q180","q181","q182","q183","q184","q185","q186","q187","q188","q189","q190","q191","q192","q193","q194","q195","q196","q197","q198","q199","q200"
+
+ ''',
+ 'scans/Manual/MultiMarkedFiles.csv': '''
+ "file_id","input_path","output_path","score","Roll_no","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22","q23","q24","q25","q26","q27","q28","q29","q30","q31","q32","q33","q34","q35","q36","q37","q38","q39","q40","q41","q42","q43","q44","q45","q46","q47","q48","q49","q50","q51","q52","q53","q54","q55","q56","q57","q58","q59","q60","q61","q62","q63","q64","q65","q66","q67","q68","q69","q70","q71","q72","q73","q74","q75","q76","q77","q78","q79","q80","q81","q82","q83","q84","q85","q86","q87","q88","q89","q90","q91","q92","q93","q94","q95","q96","q97","q98","q99","q100","q101","q102","q103","q104","q105","q106","q107","q108","q109","q110","q111","q112","q113","q114","q115","q116","q117","q118","q119","q120","q121","q122","q123","q124","q125","q126","q127","q128","q129","q130","q131","q132","q133","q134","q135","q136","q137","q138","q139","q140","q141","q142","q143","q144","q145","q146","q147","q148","q149","q150","q151","q152","q153","q154","q155","q156","q157","q158","q159","q160","q161","q162","q163","q164","q165","q166","q167","q168","q169","q170","q171","q172","q173","q174","q175","q176","q177","q178","q179","q180","q181","q182","q183","q184","q185","q186","q187","q188","q189","q190","q191","q192","q193","q194","q195","q196","q197","q198","q199","q200"
+
+ ''',
+ 'scans/Results/Results_05AM.csv': '''
+ "file_id","input_path","output_path","score","Roll_no","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22","q23","q24","q25","q26","q27","q28","q29","q30","q31","q32","q33","q34","q35","q36","q37","q38","q39","q40","q41","q42","q43","q44","q45","q46","q47","q48","q49","q50","q51","q52","q53","q54","q55","q56","q57","q58","q59","q60","q61","q62","q63","q64","q65","q66","q67","q68","q69","q70","q71","q72","q73","q74","q75","q76","q77","q78","q79","q80","q81","q82","q83","q84","q85","q86","q87","q88","q89","q90","q91","q92","q93","q94","q95","q96","q97","q98","q99","q100","q101","q102","q103","q104","q105","q106","q107","q108","q109","q110","q111","q112","q113","q114","q115","q116","q117","q118","q119","q120","q121","q122","q123","q124","q125","q126","q127","q128","q129","q130","q131","q132","q133","q134","q135","q136","q137","q138","q139","q140","q141","q142","q143","q144","q145","q146","q147","q148","q149","q150","q151","q152","q153","q154","q155","q156","q157","q158","q159","q160","q161","q162","q163","q164","q165","q166","q167","q168","q169","q170","q171","q172","q173","q174","q175","q176","q177","q178","q179","q180","q181","q182","q183","q184","q185","q186","q187","q188","q189","q190","q191","q192","q193","q194","q195","q196","q197","q198","q199","q200"
+ "scan-type-1.jpg","samples/community/UmarFarootAPS/scans/scan-type-1.jpg","outputs/community/UmarFarootAPS/scans/CheckedOMRs/scan-type-1.jpg","49.0","2468","A","C","B","C","A","D","B","C","B","D","C","A","C","D","B","C","A","B","C","A","C","B","D","C","A","B","D","C","A","C","B","D","B","A","C","D","B","C","A","C","D","A","C","D","A","B","D","C","A","C","D","B","C","A","C","D","B","C","D","A","B","C","B","C","D","B","D","A","C","B","D","A","B","C","B","A","C","D","B","A","C","B","C","B","A","D","B","A","C","D","B","D","B","C","B","D","A","C","B","C","B","C","D","B","C","A","B","C","A","D","C","B","D","B","A","B","C","D","D","C","B","A","B","C","D","C","B","A","B","C","D","C","B","A","B","C","D","C","B","A","B","C","B","A","C","B","A","C","A","B","C","B","C","B","A","C","A","C","B","B","C","B","A","C","A","B","A","B","A","B","C","D","B","C","A","C","D","C","A","C","B","A","C","A","B","C","B","D","A","B","C","D","C","B","B","C","A","B","C","B"
+ "scan-type-2.jpg","samples/community/UmarFarootAPS/scans/scan-type-2.jpg","outputs/community/UmarFarootAPS/scans/CheckedOMRs/scan-type-2.jpg","20.0","0234","A","B","C","D","C","B","A","B","C","D","C","B","A","B","C","D","C","B","A","B","C","D","C","B","A","B","C","D","C","B","A","B","C","D","C","B","A","B","C","D","C","B","A","B","C","D","C","B","A","B","A","D","","","AD","","","","A","D","","","","","","","D","A","","D","","A","","D","","","","A","","","C","","","D","","","A","","","","D","","C","","A","","C","","D","B","B","","","A","","D","","","","D","","","","","A","D","","","B","","","D","","","A","","","D","","","","","","D","","","","A","D","","","A","","B","","D","","","","C","C","D","D","A","","D","","A","D","","","D","","B","D","","","D","","D","B","","","","D","","A","","","","D","","B","","","","","","D","","","A","","","A","","D","","","D"
+
+ ''',
+ })
+# ---
+# name: test_run_community_ibrahimkilic
+ dict({
+ 'Manual/ErrorFiles.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5"
+
+ ''',
+ 'Manual/MultiMarkedFiles.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5"
+
+ ''',
+ 'Results/Results_05AM.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5"
+ "yes_no_questionnarie.jpg","samples/community/ibrahimkilic/yes_no_questionnarie.jpg","outputs/community/ibrahimkilic/CheckedOMRs/yes_no_questionnarie.jpg","0","no","no","no","no","no"
+
+ ''',
+ })
+# ---
+# name: test_run_sample1
+ dict({
+ 'MobileCamera/Manual/ErrorFiles.csv': '''
+ "file_id","input_path","output_path","score","Roll","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20"
+
+ ''',
+ 'MobileCamera/Manual/MultiMarkedFiles.csv': '''
+ "file_id","input_path","output_path","score","Roll","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20"
+
+ ''',
+ 'MobileCamera/Results/Results_05AM.csv': '''
+ "file_id","input_path","output_path","score","Roll","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20"
+ "sheet1.jpg","samples/sample1/MobileCamera/sheet1.jpg","outputs/sample1/MobileCamera/CheckedOMRs/sheet1.jpg","0","E503110026","B","","D","B","6","11","20","7","16","B","D","C","D","A","D","B","A","C","C","D"
+
+ ''',
+ })
+# ---
+# name: test_run_sample2
+ dict({
+ 'AdrianSample/Manual/ErrorFiles.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5"
+
+ ''',
+ 'AdrianSample/Manual/MultiMarkedFiles.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5"
+
+ ''',
+ 'AdrianSample/Results/Results_05AM.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5"
+ "adrian_omr.png","samples/sample2/AdrianSample/adrian_omr.png","outputs/sample2/AdrianSample/CheckedOMRs/adrian_omr.png","0","B","E","A","C","B"
+ "adrian_omr_2.png","samples/sample2/AdrianSample/adrian_omr_2.png","outputs/sample2/AdrianSample/CheckedOMRs/adrian_omr_2.png","0","C","E","A","B","B"
+
+ ''',
+ })
+# ---
+# name: test_run_sample3
+ dict({
+ 'colored-thick-sheet/Manual/ErrorFiles.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22","q23","q24","q25","q26","q27","q28","q29","q30","q31","q32","q33","q34","q35","q36","q37","q38","q39","q40","q41","q42","q43","q44","q45","q46","q47","q48","q49","q50","q51","q52","q53","q54","q55","q56","q57","q58","q59","q60","q61","q62","q63","q64","q65","q66","q67","q68","q69","q70","q71","q72","q73","q74","q75","q76","q77","q78","q79","q80","q81","q82","q83","q84","q85","q86","q87","q88","q89","q90","q91","q92","q93","q94","q95","q96","q97","q98","q99","q100"
+
+ ''',
+ 'colored-thick-sheet/Manual/MultiMarkedFiles.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22","q23","q24","q25","q26","q27","q28","q29","q30","q31","q32","q33","q34","q35","q36","q37","q38","q39","q40","q41","q42","q43","q44","q45","q46","q47","q48","q49","q50","q51","q52","q53","q54","q55","q56","q57","q58","q59","q60","q61","q62","q63","q64","q65","q66","q67","q68","q69","q70","q71","q72","q73","q74","q75","q76","q77","q78","q79","q80","q81","q82","q83","q84","q85","q86","q87","q88","q89","q90","q91","q92","q93","q94","q95","q96","q97","q98","q99","q100"
+
+ ''',
+ 'colored-thick-sheet/Results/Results_05AM.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22","q23","q24","q25","q26","q27","q28","q29","q30","q31","q32","q33","q34","q35","q36","q37","q38","q39","q40","q41","q42","q43","q44","q45","q46","q47","q48","q49","q50","q51","q52","q53","q54","q55","q56","q57","q58","q59","q60","q61","q62","q63","q64","q65","q66","q67","q68","q69","q70","q71","q72","q73","q74","q75","q76","q77","q78","q79","q80","q81","q82","q83","q84","q85","q86","q87","q88","q89","q90","q91","q92","q93","q94","q95","q96","q97","q98","q99","q100"
+ "rgb-100-gsm.jpg","samples/sample3/colored-thick-sheet/rgb-100-gsm.jpg","outputs/sample3/colored-thick-sheet/CheckedOMRs/rgb-100-gsm.jpg","0","D","D","A","","C","C","B","","A","C","C","D","A","D","A","C","A","D","B","D","D","C","D","D","D","D","","B","A","D","D","C","","B","","C","D","","","A","","A","C","C","B","C","A","A","C","","C","","D","B","C","","B","C","D","","","C","C","","C","A","B","C","","","","","D","D","C","D","A","","","B","","B","D","C","C","","D","","D","C","D","A","","A","","","A","C","B","A"
+
+ ''',
+ 'xeroxed-thin-sheet/Manual/ErrorFiles.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22","q23","q24","q25","q26","q27","q28","q29","q30","q31","q32","q33","q34","q35","q36","q37","q38","q39","q40","q41","q42","q43","q44","q45","q46","q47","q48","q49","q50","q51","q52","q53","q54","q55","q56","q57","q58","q59","q60","q61","q62","q63","q64","q65","q66","q67","q68","q69","q70","q71","q72","q73","q74","q75","q76","q77","q78","q79","q80","q81","q82","q83","q84","q85","q86","q87","q88","q89","q90","q91","q92","q93","q94","q95","q96","q97","q98","q99","q100"
+
+ ''',
+ 'xeroxed-thin-sheet/Manual/MultiMarkedFiles.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22","q23","q24","q25","q26","q27","q28","q29","q30","q31","q32","q33","q34","q35","q36","q37","q38","q39","q40","q41","q42","q43","q44","q45","q46","q47","q48","q49","q50","q51","q52","q53","q54","q55","q56","q57","q58","q59","q60","q61","q62","q63","q64","q65","q66","q67","q68","q69","q70","q71","q72","q73","q74","q75","q76","q77","q78","q79","q80","q81","q82","q83","q84","q85","q86","q87","q88","q89","q90","q91","q92","q93","q94","q95","q96","q97","q98","q99","q100"
+
+ ''',
+ 'xeroxed-thin-sheet/Results/Results_05AM.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22","q23","q24","q25","q26","q27","q28","q29","q30","q31","q32","q33","q34","q35","q36","q37","q38","q39","q40","q41","q42","q43","q44","q45","q46","q47","q48","q49","q50","q51","q52","q53","q54","q55","q56","q57","q58","q59","q60","q61","q62","q63","q64","q65","q66","q67","q68","q69","q70","q71","q72","q73","q74","q75","q76","q77","q78","q79","q80","q81","q82","q83","q84","q85","q86","q87","q88","q89","q90","q91","q92","q93","q94","q95","q96","q97","q98","q99","q100"
+ "grayscale-80-gsm.jpg","samples/sample3/xeroxed-thin-sheet/grayscale-80-gsm.jpg","outputs/sample3/xeroxed-thin-sheet/CheckedOMRs/grayscale-80-gsm.jpg","0","C","D","A","C","C","C","B","A","C","C","B","D","B","D","C","C","B","D","B","D","C","C","C","B","D","D","D","B","A","D","D","C","A","B","C","A","D","A","A","A","D","D","B","A","B","C","B","A","C","D","C","D","A","B","C","A","C","C","C","D","B","C","C","C","C","A","D","A","D","A","D","C","C","D","C","D","A","A","C","B","C","D","C","A","B","C","B","D","A","A","C","A","B","D","C","D","A","C","B","A"
+
+ ''',
+ })
+# ---
+# name: test_run_sample4
+ dict({
+ 'Manual/ErrorFiles.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11"
+
+ ''',
+ 'Manual/MultiMarkedFiles.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11"
+
+ ''',
+ 'Results/Results_05AM.csv': '''
+ "file_id","input_path","output_path","score","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11"
+ "IMG_20201116_143512.jpg","samples/sample4/IMG_20201116_143512.jpg","outputs/sample4/CheckedOMRs/IMG_20201116_143512.jpg","33.0","B","D","C","B","D","C","BC","A","C","D","C"
+ "IMG_20201116_150717658.jpg","samples/sample4/IMG_20201116_150717658.jpg","outputs/sample4/CheckedOMRs/IMG_20201116_150717658.jpg","33.0","B","D","C","B","D","C","BC","A","C","D","C"
+ "IMG_20201116_150750830.jpg","samples/sample4/IMG_20201116_150750830.jpg","outputs/sample4/CheckedOMRs/IMG_20201116_150750830.jpg","-2.0","A","","D","C","AC","A","D","B","C","D","D"
+
+ ''',
+ })
+# ---
+# name: test_run_sample5
+ dict({
+ 'ScanBatch1/Manual/ErrorFiles.csv': '''
+ "file_id","input_path","output_path","score","Roll","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22"
+
+ ''',
+ 'ScanBatch1/Manual/MultiMarkedFiles.csv': '''
+ "file_id","input_path","output_path","score","Roll","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22"
+
+ ''',
+ 'ScanBatch1/Results/Results_05AM.csv': '''
+ "file_id","input_path","output_path","score","Roll","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22"
+ "camscanner-1.jpg","samples/sample5/ScanBatch1/camscanner-1.jpg","outputs/sample5/ScanBatch1/CheckedOMRs/camscanner-1.jpg","-4.0","E204420102","D","C","A","C","B","08","52","21","85","36","B","C","A","A","D","C","C","AD","A","A","D",""
+
+ ''',
+ 'ScanBatch2/Manual/ErrorFiles.csv': '''
+ "file_id","input_path","output_path","score","Roll","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22"
+
+ ''',
+ 'ScanBatch2/Manual/MultiMarkedFiles.csv': '''
+ "file_id","input_path","output_path","score","Roll","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22"
+
+ ''',
+ 'ScanBatch2/Results/Results_05AM.csv': '''
+ "file_id","input_path","output_path","score","Roll","q1","q2","q3","q4","q5","q6","q7","q8","q9","q10","q11","q12","q13","q14","q15","q16","q17","q18","q19","q20","q21","q22"
+ "camscanner-2.jpg","samples/sample5/ScanBatch2/camscanner-2.jpg","outputs/sample5/ScanBatch2/CheckedOMRs/camscanner-2.jpg","55.0","E204420109","C","C","B","C","C","01","19","10","10","18","D","A","D","D","D","C","C","C","C","D","B","A"
+
+ ''',
+ })
+# ---
+# name: test_run_sample6
+ dict({
+ 'Manual/ErrorFiles.csv': '''
+ "file_id","input_path","output_path","score","Roll"
+
+ ''',
+ 'Manual/MultiMarkedFiles.csv': '''
+ "file_id","input_path","output_path","score","Roll"
+
+ ''',
+ 'Results/Results_05AM.csv': '''
+ "file_id","input_path","output_path","score","Roll"
+ "reference.png","samples/sample6/reference.png","outputs/sample6/CheckedOMRs/reference.png","0","A"
+
+ ''',
+ 'doc-scans/Manual/ErrorFiles.csv': '''
+ "file_id","input_path","output_path","score","Roll"
+
+ ''',
+ 'doc-scans/Manual/MultiMarkedFiles.csv': '''
+ "file_id","input_path","output_path","score","Roll"
+
+ ''',
+ 'doc-scans/Results/Results_05AM.csv': '''
+ "file_id","input_path","output_path","score","Roll"
+ "sample_roll_01.jpg","samples/sample6/doc-scans/sample_roll_01.jpg","outputs/sample6/doc-scans/CheckedOMRs/sample_roll_01.jpg","0","A0188877Y"
+ "sample_roll_02.jpg","samples/sample6/doc-scans/sample_roll_02.jpg","outputs/sample6/doc-scans/CheckedOMRs/sample_roll_02.jpg","0","A0203959W"
+ "sample_roll_03.jpg","samples/sample6/doc-scans/sample_roll_03.jpg","outputs/sample6/doc-scans/CheckedOMRs/sample_roll_03.jpg","0","A0204729A"
+
+ ''',
+ })
+# ---
diff --git a/src/tests/test_all_samples.py b/src/tests/test_all_samples.py
new file mode 100644
index 00000000..9efe94ff
--- /dev/null
+++ b/src/tests/test_all_samples.py
@@ -0,0 +1,112 @@
+import os
+import shutil
+from glob import glob
+
+from src.tests.utils import run_entry_point, setup_mocker_patches
+
+
+def read_file(path):
+ with open(path) as file:
+ return file.read()
+
+
+def run_sample(mocker, sample_path):
+ setup_mocker_patches(mocker)
+
+ input_path = os.path.join("samples", sample_path)
+ output_dir = os.path.join("outputs", sample_path)
+ if os.path.exists(output_dir):
+ print(
+ f"Warning: output directory already exists: {output_dir}. This may affect the test execution."
+ )
+
+ run_entry_point(input_path, output_dir)
+
+ sample_outputs = extract_sample_outputs(output_dir)
+
+ print(f"Note: removing output directory: {output_dir}")
+ shutil.rmtree(output_dir)
+
+ return sample_outputs
+
+
+EXT = "*.csv"
+
+
+def extract_sample_outputs(output_dir):
+ sample_outputs = {}
+ for _dir, _subdir, _files in os.walk(output_dir):
+ for file in glob(os.path.join(_dir, EXT)):
+ relative_path = os.path.relpath(file, output_dir)
+ sample_outputs[relative_path] = read_file(file)
+ return sample_outputs
+
+
+def test_run_answer_key_using_csv(mocker, snapshot):
+ sample_outputs = run_sample(mocker, "answer-key/using-csv")
+ assert snapshot == sample_outputs
+
+
+def test_run_answer_key_weighted_answers(mocker, snapshot):
+ sample_outputs = run_sample(mocker, "answer-key/weighted-answers")
+ assert snapshot == sample_outputs
+
+
+def test_run_sample1(mocker, snapshot):
+ sample_outputs = run_sample(mocker, "sample1")
+ assert snapshot == sample_outputs
+
+
+def test_run_sample2(mocker, snapshot):
+ sample_outputs = run_sample(mocker, "sample2")
+ assert snapshot == sample_outputs
+
+
+def test_run_sample3(mocker, snapshot):
+ sample_outputs = run_sample(mocker, "sample3")
+ assert snapshot == sample_outputs
+
+
+def test_run_sample4(mocker, snapshot):
+ sample_outputs = run_sample(mocker, "sample4")
+ assert snapshot == sample_outputs
+
+
+def test_run_sample5(mocker, snapshot):
+ sample_outputs = run_sample(mocker, "sample5")
+ assert snapshot == sample_outputs
+
+
+def test_run_sample6(mocker, snapshot):
+ sample_outputs = run_sample(mocker, "sample6")
+ assert snapshot == sample_outputs
+
+
+def test_run_community_Antibodyy(mocker, snapshot):
+ sample_outputs = run_sample(mocker, "community/Antibodyy")
+ assert snapshot == sample_outputs
+
+
+def test_run_community_ibrahimkilic(mocker, snapshot):
+ sample_outputs = run_sample(mocker, "community/ibrahimkilic")
+ assert snapshot == sample_outputs
+
+
+def test_run_community_Sandeep_1507(mocker, snapshot):
+ sample_outputs = run_sample(mocker, "community/Sandeep-1507")
+ assert snapshot == sample_outputs
+
+
+def test_run_community_Shamanth(mocker, snapshot):
+ sample_outputs = run_sample(mocker, "community/Shamanth")
+ assert snapshot == sample_outputs
+
+
+def test_run_community_UmarFarootAPS(mocker, snapshot):
+ sample_outputs = run_sample(mocker, "community/UmarFarootAPS")
+ assert snapshot == sample_outputs
+
+
+def test_run_community_UPSC_mock(mocker, snapshot):
+ sample_outputs = run_sample(mocker, "community/UPSC-mock")
+ assert snapshot == sample_outputs
diff --git a/src/tests/test_edge_cases.py b/src/tests/test_edge_cases.py
new file mode 100644
index 00000000..f3d6fd45
--- /dev/null
+++ b/src/tests/test_edge_cases.py
@@ -0,0 +1,95 @@
+import os
+from pathlib import Path
+
+import pandas as pd
+
+from src.tests.test_samples.sample2.boilerplate import (
+ CONFIG_BOILERPLATE,
+ TEMPLATE_BOILERPLATE,
+)
+from src.tests.utils import (
+ generate_write_jsons_and_run,
+ remove_file,
+ run_entry_point,
+ setup_mocker_patches,
+)
+
+FROZEN_TIMESTAMP = "1970-01-01"
+CURRENT_DIR = Path("src/tests")
+BASE_SAMPLE_PATH = CURRENT_DIR.joinpath("test_samples", "sample2")
+BASE_RESULTS_CSV_PATH = os.path.join(
+ "outputs", BASE_SAMPLE_PATH, "Results", "Results_05AM.csv"
+)
+BASE_MULTIMARKED_CSV_PATH = os.path.join(
+ "outputs", BASE_SAMPLE_PATH, "Manual", "MultiMarkedFiles.csv"
+)
+
+
+def run_sample(mocker, input_path):
+ setup_mocker_patches(mocker)
+ output_dir = os.path.join("outputs", input_path)
+ run_entry_point(input_path, output_dir)
+
+
+def extract_output_data(path):
+ output_data = pd.read_csv(path, keep_default_na=False)
+ return output_data
+
+
+write_jsons_and_run = generate_write_jsons_and_run(
+ run_sample,
+ sample_path=BASE_SAMPLE_PATH,
+ template_boilerplate=TEMPLATE_BOILERPLATE,
+ config_boilerplate=CONFIG_BOILERPLATE,
+)
+
+
+def test_config_low_dimensions(mocker):
+ def modify_config(config):
+ config["dimensions"]["processing_height"] = 1000
+ config["dimensions"]["processing_width"] = 1000
+
+ exception = write_jsons_and_run(mocker, modify_config=modify_config)
+
+ assert str(exception) == "No Error"
+
+
+def test_different_bubble_dimensions(mocker):
+ # Prevent appending to output csv:
+ remove_file(BASE_RESULTS_CSV_PATH)
+ remove_file(BASE_MULTIMARKED_CSV_PATH)
+
+ exception = write_jsons_and_run(mocker)
+ assert str(exception) == "No Error"
+ original_output_data = extract_output_data(BASE_RESULTS_CSV_PATH)
+
+ def modify_template(template):
+ # Incorrect global bubble size
+ template["bubbleDimensions"] = [5, 5]
+ # Correct bubble size for MCQBlock1a1
+ template["fieldBlocks"]["MCQBlock1a1"]["bubbleDimensions"] = [32, 32]
+ # Incorrect bubble size for MCQBlock1a11
+ template["fieldBlocks"]["MCQBlock1a11"]["bubbleDimensions"] = [10, 10]
+
+ remove_file(BASE_RESULTS_CSV_PATH)
+ remove_file(BASE_MULTIMARKED_CSV_PATH)
+ exception = write_jsons_and_run(mocker, modify_template=modify_template)
+ assert str(exception) == "No Error"
+
+ results_output_data = extract_output_data(BASE_RESULTS_CSV_PATH)
+
+ assert results_output_data.empty
+
+ output_data = extract_output_data(BASE_MULTIMARKED_CSV_PATH)
+
+ equal_columns = [f"q{i}" for i in range(1, 18)]
+ assert (
+ output_data[equal_columns].iloc[0].to_list()
+ == original_output_data[equal_columns].iloc[0].to_list()
+ )
+
+ unequal_columns = [f"q{i}" for i in range(168, 185)]
+ assert not (
+ output_data[unequal_columns].iloc[0].to_list()
+ == original_output_data[unequal_columns].iloc[0].to_list()
+ )
diff --git a/src/tests/test_samples/sample1/boilerplate.py b/src/tests/test_samples/sample1/boilerplate.py
new file mode 100644
index 00000000..74dda465
--- /dev/null
+++ b/src/tests/test_samples/sample1/boilerplate.py
@@ -0,0 +1,14 @@
+TEMPLATE_BOILERPLATE = {
+ "pageDimensions": [300, 400],
+ "bubbleDimensions": [25, 25],
+ "preProcessors": [{"name": "CropPage", "options": {"morphKernel": [10, 10]}}],
+ "fieldBlocks": {
+ "MCQ_Block_1": {
+ "fieldType": "QTYPE_MCQ5",
+ "origin": [65, 60],
+ "fieldLabels": ["q1..5"],
+ "labelsGap": 52,
+ "bubblesGap": 41,
+ }
+ },
+}
diff --git a/src/tests/test_samples/sample1/sample.png b/src/tests/test_samples/sample1/sample.png
new file mode 100644
index 00000000..d8db0994
Binary files /dev/null and b/src/tests/test_samples/sample1/sample.png differ
diff --git a/src/tests/test_samples/sample2/boilerplate.py b/src/tests/test_samples/sample2/boilerplate.py
new file mode 100644
index 00000000..f43c3093
--- /dev/null
+++ b/src/tests/test_samples/sample2/boilerplate.py
@@ -0,0 +1,39 @@
+TEMPLATE_BOILERPLATE = {
+ "pageDimensions": [2550, 3300],
+ "bubbleDimensions": [32, 32],
+ "preProcessors": [
+ {
+ "name": "CropOnMarkers",
+ "options": {
+ "relativePath": "omr_marker.jpg",
+ "sheetToMarkerWidthRatio": 17,
+ },
+ }
+ ],
+ "fieldBlocks": {
+ "MCQBlock1a1": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [197, 300],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": ["q1..17"],
+ },
+ "MCQBlock1a11": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [1770, 1310],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": ["q168..184"],
+ },
+ },
+}
+
+CONFIG_BOILERPLATE = {
+ "dimensions": {
+ "display_height": 960,
+ "display_width": 1280,
+ "processing_height": 1640,
+ "processing_width": 1332,
+ },
+ "outputs": {"show_image_level": 0, "filter_out_multimarked_files": True},
+}
diff --git a/src/tests/test_samples/sample2/omr_marker.jpg b/src/tests/test_samples/sample2/omr_marker.jpg
new file mode 100644
index 00000000..0929feec
Binary files /dev/null and b/src/tests/test_samples/sample2/omr_marker.jpg differ
diff --git a/src/tests/test_samples/sample2/sample.jpg b/src/tests/test_samples/sample2/sample.jpg
new file mode 100644
index 00000000..2eef7c32
Binary files /dev/null and b/src/tests/test_samples/sample2/sample.jpg differ
diff --git a/src/tests/test_template_validations.py b/src/tests/test_template_validations.py
new file mode 100644
index 00000000..bd321a72
--- /dev/null
+++ b/src/tests/test_template_validations.py
@@ -0,0 +1,159 @@
+import os
+from pathlib import Path
+
+from src.tests.test_samples.sample1.boilerplate import TEMPLATE_BOILERPLATE
+from src.tests.utils import (
+ generate_write_jsons_and_run,
+ run_entry_point,
+ setup_mocker_patches,
+)
+
+FROZEN_TIMESTAMP = "1970-01-01"
+CURRENT_DIR = Path("src/tests")
+BASE_SAMPLE_PATH = CURRENT_DIR.joinpath("test_samples", "sample1")
+BASE_SAMPLE_TEMPLATE_PATH = BASE_SAMPLE_PATH.joinpath("template.json")
+
+
+def run_sample(mocker, input_path):
+ setup_mocker_patches(mocker)
+ output_dir = os.path.join("outputs", input_path)
+ run_entry_point(input_path, output_dir)
+
+
+write_jsons_and_run = generate_write_jsons_and_run(
+ run_sample,
+ sample_path=BASE_SAMPLE_PATH,
+ template_boilerplate=TEMPLATE_BOILERPLATE,
+)
+
+
+def test_no_input_dir(mocker):
+ try:
+ run_sample(mocker, "X")
+ except Exception as e:
+ assert str(e) == "Given input directory does not exist: 'X'"
+
+
+def test_no_template(mocker):
+ if os.path.exists(BASE_SAMPLE_TEMPLATE_PATH):
+ os.remove(BASE_SAMPLE_TEMPLATE_PATH)
+ try:
+ run_sample(mocker, BASE_SAMPLE_PATH)
+ except Exception as e:
+ assert (
+ str(e)
+ == "No template file found in the directory tree of src/tests/test_samples/sample1"
+ )
+
+
+def test_empty_template(mocker):
+ def modify_template(_):
+ return {}
+
+ exception = write_jsons_and_run(mocker, modify_template=modify_template)
+ assert (
+ str(exception)
+ == f"Provided Template JSON is Invalid: '{BASE_SAMPLE_TEMPLATE_PATH}'"
+ )
+
+
+def test_invalid_field_type(mocker):
+ def modify_template(template):
+ template["fieldBlocks"]["MCQ_Block_1"]["fieldType"] = "X"
+
+ exception = write_jsons_and_run(mocker, modify_template=modify_template)
+ assert (
+ str(exception)
+ == f"Provided Template JSON is Invalid: '{BASE_SAMPLE_TEMPLATE_PATH}'"
+ )
+
+
+def test_overflow_labels(mocker):
+ def modify_template(template):
+ template["fieldBlocks"]["MCQ_Block_1"]["fieldLabels"] = ["q1..100"]
+
+ exception = write_jsons_and_run(mocker, modify_template=modify_template)
+ assert (
+ str(exception)
+ == "Overflowing field block 'MCQ_Block_1' with origin [65, 60] and dimensions [189, 5173] in template with dimensions [300, 400]"
+ )
+
+
+def test_overflow_safe_dimensions(mocker):
+ def modify_template(template):
+ template["pageDimensions"] = [255, 400]
+
+ exception = write_jsons_and_run(mocker, modify_template=modify_template)
+ assert str(exception) == "No Error"
+
+
+def test_field_strings_overlap(mocker):
+ def modify_template(template):
+ template["fieldBlocks"] = {
+ **template["fieldBlocks"],
+ "New_Block": {
+ **template["fieldBlocks"]["MCQ_Block_1"],
+ "fieldLabels": ["q5"],
+ },
+ }
+
+ exception = write_jsons_and_run(mocker, modify_template=modify_template)
+ assert str(exception) == (
+ "The field strings for field block New_Block overlap with other existing fields"
+ )
+
+
+def test_custom_label_strings_overlap_single(mocker):
+ def modify_template(template):
+ template["customLabels"] = {
+ "label1": ["q1..2", "q2..3"],
+ }
+
+ exception = write_jsons_and_run(mocker, modify_template=modify_template)
+ assert (
+ str(exception)
+ == "Given field string 'q2..3' has overlapping field(s) with other fields in 'Custom Label: label1': ['q1..2', 'q2..3']"
+ )
+
+
+def test_custom_label_strings_overlap_multiple(mocker):
+ def modify_template(template):
+ template["customLabels"] = {
+ "label1": ["q1..2"],
+ "label2": ["q2..3"],
+ }
+
+ exception = write_jsons_and_run(mocker, modify_template=modify_template)
+ assert (
+ str(exception)
+ == "The field strings for custom label 'label2' overlap with other existing custom labels"
+ )
+
+
+def test_missing_field_block_labels(mocker):
+ def modify_template(template):
+ template["customLabels"] = {"Combined": ["qX", "qY"]}
+
+ exception = write_jsons_and_run(mocker, modify_template=modify_template)
+ assert (
+ str(exception)
+ == "Missing field block label(s) in the given template for ['qX', 'qY'] from 'Combined'"
+ )
+
+
+def test_missing_output_columns(mocker):
+ def modify_template(template):
+ template["outputColumns"] = ["qX", "q1..5"]
+
+ exception = write_jsons_and_run(mocker, modify_template=modify_template)
+ assert str(exception) == (
+ "Some columns are missing in the field blocks for the given output columns"
+ )
+
+
+def test_safe_missing_label_columns(mocker):
+ def modify_template(template):
+ template["outputColumns"] = ["q1..4"]
+
+ exception = write_jsons_and_run(mocker, modify_template=modify_template)
+ assert str(exception) == "No Error"
diff --git a/src/tests/utils.py b/src/tests/utils.py
new file mode 100644
index 00000000..f8e5bfab
--- /dev/null
+++ b/src/tests/utils.py
@@ -0,0 +1,97 @@
+import json
+import os
+from copy import deepcopy
+
+from freezegun import freeze_time
+
+from main import entry_point_for_args
+
+FROZEN_TIMESTAMP = "1970-01-01"
+
+
+def setup_mocker_patches(mocker):
+ mock_imshow = mocker.patch("cv2.imshow")
+ mock_imshow.return_value = True
+
+ mock_destroy_all_windows = mocker.patch("cv2.destroyAllWindows")
+ mock_destroy_all_windows.return_value = True
+
+ mock_wait_key = mocker.patch("cv2.waitKey")
+ mock_wait_key.return_value = ord("q")
+
+
+def run_entry_point(input_path, output_dir):
+ args = {
+ "autoAlign": False,
+ "debug": False,
+ "input_paths": [input_path],
+ "output_dir": output_dir,
+ "setLayout": False,
+ "silent": True,
+ }
+ with freeze_time(FROZEN_TIMESTAMP):
+ entry_point_for_args(args)
+
+
+def write_modified(modify_content, boilerplate, sample_json_path):
+ if boilerplate is None:
+ return
+
+ content = deepcopy(boilerplate)
+
+ if modify_content is not None:
+ returned_value = modify_content(content)
+ if returned_value is not None:
+ content = returned_value
+
+ with open(sample_json_path, "w") as f:
+ json.dump(content, f)
+
+
+def remove_file(path):
+ if os.path.exists(path):
+ os.remove(path)
+
+
+def generate_write_jsons_and_run(
+ run_sample,
+ sample_path,
+ template_boilerplate=None,
+ config_boilerplate=None,
+ evaluation_boilerplate=None,
+):
+ if (template_boilerplate or config_boilerplate or evaluation_boilerplate) is None:
+ raise Exception(
+ f"No boilerplates found. Provide atleast one boilerplate to write json."
+ )
+
+ def write_jsons_and_run(
+ mocker,
+ modify_template=None,
+ modify_config=None,
+ modify_evaluation=None,
+ ):
+ sample_template_path, sample_config_path, sample_evaluation_path = (
+ sample_path.joinpath("template.json"),
+ sample_path.joinpath("config.json"),
+ sample_path.joinpath("evaluation.json"),
+ )
+ write_modified(modify_template, template_boilerplate, sample_template_path)
+ write_modified(modify_config, config_boilerplate, sample_config_path)
+ write_modified(
+ modify_evaluation, evaluation_boilerplate, sample_evaluation_path
+ )
+
+ exception = "No Error"
+ try:
+ run_sample(mocker, sample_path)
+ except Exception as e:
+ exception = e
+
+ remove_file(sample_template_path)
+ remove_file(sample_config_path)
+ remove_file(sample_evaluation_path)
+
+ return exception
+
+ return write_jsons_and_run
diff --git a/src/utils/__init__.py b/src/utils/__init__.py
new file mode 100644
index 00000000..e69de29b
diff --git a/src/utils/file.py b/src/utils/file.py
new file mode 100644
index 00000000..08536ff1
--- /dev/null
+++ b/src/utils/file.py
@@ -0,0 +1,95 @@
+import argparse
+import json
+import os
+from csv import QUOTE_NONNUMERIC
+from time import localtime, strftime
+
+import pandas as pd
+
+from src.logger import logger
+
+
+def load_json(path, **rest):
+ try:
+ with open(path, "r") as f:
+ loaded = json.load(f, **rest)
+ except json.decoder.JSONDecodeError as error:
+ logger.critical(f"Error when loading json file at: '{path}'\n{error}")
+ exit(1)
+ return loaded
+
+
+class Paths:
+ def __init__(self, output_dir):
+ self.output_dir = output_dir
+ self.save_marked_dir = output_dir.joinpath("CheckedOMRs")
+ self.results_dir = output_dir.joinpath("Results")
+ self.manual_dir = output_dir.joinpath("Manual")
+ self.evaluation_dir = output_dir.joinpath("Evaluation")
+ self.errors_dir = self.manual_dir.joinpath("ErrorFiles")
+ self.multi_marked_dir = self.manual_dir.joinpath("MultiMarkedFiles")
+
+
+def setup_dirs_for_paths(paths):
+ logger.info("Checking Directories...")
+ for save_output_dir in [paths.save_marked_dir]:
+ if not os.path.exists(save_output_dir):
+ logger.info(f"Created : {save_output_dir}")
+ os.makedirs(save_output_dir)
+ os.mkdir(save_output_dir.joinpath("stack"))
+ os.mkdir(save_output_dir.joinpath("_MULTI_"))
+ os.mkdir(save_output_dir.joinpath("_MULTI_", "stack"))
+
+ for save_output_dir in [paths.manual_dir, paths.results_dir, paths.evaluation_dir]:
+ if not os.path.exists(save_output_dir):
+ logger.info(f"Created : {save_output_dir}")
+ os.makedirs(save_output_dir)
+
+ for save_output_dir in [paths.multi_marked_dir, paths.errors_dir]:
+ if not os.path.exists(save_output_dir):
+ logger.info(f"Created : {save_output_dir}")
+ os.makedirs(save_output_dir)
+
+
+def setup_outputs_for_template(paths, template):
+ # TODO: consider moving this into a class instance
+ ns = argparse.Namespace()
+ logger.info("Checking Files...")
+
+ # Include current output paths
+ ns.paths = paths
+
+ ns.empty_resp = [""] * len(template.output_columns)
+ ns.sheetCols = [
+ "file_id",
+ "input_path",
+ "output_path",
+ "score",
+ ] + template.output_columns
+ ns.OUTPUT_SET = []
+ ns.files_obj = {}
+ TIME_NOW_HRS = strftime("%I%p", localtime())
+ ns.filesMap = {
+ "Results": os.path.join(paths.results_dir, f"Results_{TIME_NOW_HRS}.csv"),
+ "MultiMarked": os.path.join(paths.manual_dir, "MultiMarkedFiles.csv"),
+ "Errors": os.path.join(paths.manual_dir, "ErrorFiles.csv"),
+ }
+
+ for file_key, file_name in ns.filesMap.items():
+ if not os.path.exists(file_name):
+ logger.info(f"Created new file: '{file_name}'")
+ # moved handling of files to pandas csv writer
+ ns.files_obj[file_key] = file_name
+ # Create Header Columns
+ pd.DataFrame([ns.sheetCols], dtype=str).to_csv(
+ ns.files_obj[file_key],
+ mode="a",
+ quoting=QUOTE_NONNUMERIC,
+ header=False,
+ index=False,
+ )
+ else:
+ logger.info(f"Present : appending to '{file_name}'")
+ ns.files_obj[file_key] = open(file_name, "a")
+
+ return ns
diff --git a/src/utils/image.py b/src/utils/image.py
new file mode 100644
index 00000000..bba6f6d1
--- /dev/null
+++ b/src/utils/image.py
@@ -0,0 +1,155 @@
+"""
+
+ OMRChecker
+
+ Author: Udayraj Deshmukh
+ Github: https://github.com/Udayraj123
+
+"""
+import cv2
+import matplotlib.pyplot as plt
+import numpy as np
+
+from src.logger import logger
+
+plt.rcParams["figure.figsize"] = (10.0, 8.0)
+CLAHE_HELPER = cv2.createCLAHE(clipLimit=5.0, tileGridSize=(8, 8))
+
+
+class ImageUtils:
+ """A Static-only Class to hold common image processing utilities & wrappers over OpenCV functions"""
+
+ @staticmethod
+ def save_img(path, final_marked):
+ logger.info(f"Saving Image to '{path}'")
+ cv2.imwrite(path, final_marked)
+
+ @staticmethod
+ def resize_util(img, u_width, u_height=None):
+ if u_height is None:
+ h, w = img.shape[:2]
+ u_height = int(h * u_width / w)
+ return cv2.resize(img, (int(u_width), int(u_height)))
+
+ @staticmethod
+ def resize_util_h(img, u_height, u_width=None):
+ if u_width is None:
+ h, w = img.shape[:2]
+ u_width = int(w * u_height / h)
+ return cv2.resize(img, (int(u_width), int(u_height)))
+
+ @staticmethod
+ def grab_contours(cnts):
+ # source: imutils package
+
+ # if the length the contours tuple returned by cv2.findContours
+ # is '2' then we are using either OpenCV v2.4, v4-beta, or
+ # v4-official
+ if len(cnts) == 2:
+ cnts = cnts[0]
+
+ # if the length of the contours tuple is '3' then we are using
+ # either OpenCV v3, v4-pre, or v4-alpha
+ elif len(cnts) == 3:
+ cnts = cnts[1]
+
+ # otherwise OpenCV has changed their cv2.findContours return
+ # signature yet again and I have no idea WTH is going on
+ else:
+ raise Exception(
+ (
+ "Contours tuple must have length 2 or 3, "
+ "otherwise OpenCV changed their cv2.findContours return "
+ "signature yet again. Refer to OpenCV's documentation "
+ "in that case"
+ )
+ )
+
+ # return the actual contours array
+ return cnts
+
+ @staticmethod
+ def normalize_util(img, alpha=0, beta=255):
+ return cv2.normalize(img, alpha, beta, norm_type=cv2.NORM_MINMAX)
+
+ @staticmethod
+ def auto_canny(image, sigma=0.93):
+ # compute the median of the single channel pixel intensities
+ v = np.median(image)
+
+ # apply automatic Canny edge detection using the computed median
+ lower = int(max(0, (1.0 - sigma) * v))
+ upper = int(min(255, (1.0 + sigma) * v))
+ edged = cv2.Canny(image, lower, upper)
+
+ # return the edged image
+ return edged
+
+ @staticmethod
+ def adjust_gamma(image, gamma=1.0):
+ # build a lookup table mapping the pixel values [0, 255] to
+ # their adjusted gamma values
+ inv_gamma = 1.0 / gamma
+ table = np.array(
+ [((i / 255.0) ** inv_gamma) * 255 for i in np.arange(0, 256)]
+ ).astype("uint8")
+
+ # apply gamma correction using the lookup table
+ return cv2.LUT(image, table)
+
+ @staticmethod
+ def four_point_transform(image, pts):
+ # obtain a consistent order of the points and unpack them
+ # individually
+ rect = ImageUtils.order_points(pts)
+ (tl, tr, br, bl) = rect
+
+ # compute the width of the new image, which will be the
+ width_a = np.sqrt(((br[0] - bl[0]) ** 2) + ((br[1] - bl[1]) ** 2))
+ width_b = np.sqrt(((tr[0] - tl[0]) ** 2) + ((tr[1] - tl[1]) ** 2))
+
+ max_width = max(int(width_a), int(width_b))
+ # max_width = max(int(np.linalg.norm(br-bl)), int(np.linalg.norm(tr-tl)))
+
+ # compute the height of the new image, which will be the
+ height_a = np.sqrt(((tr[0] - br[0]) ** 2) + ((tr[1] - br[1]) ** 2))
+ height_b = np.sqrt(((tl[0] - bl[0]) ** 2) + ((tl[1] - bl[1]) ** 2))
+ max_height = max(int(height_a), int(height_b))
+ # max_height = max(int(np.linalg.norm(tr-br)), int(np.linalg.norm(tl-br)))
+
+ # now that we have the dimensions of the new image, construct
+ # the set of destination points to obtain a "birds eye view",
+ # (i.e. top-down view) of the image, again specifying points
+ # in the top-left, top-right, bottom-right, and bottom-left
+ # order
+ dst = np.array(
+ [
+ [0, 0],
+ [max_width - 1, 0],
+ [max_width - 1, max_height - 1],
+ [0, max_height - 1],
+ ],
+ dtype="float32",
+ )
+
+ transform_matrix = cv2.getPerspectiveTransform(rect, dst)
+ warped = cv2.warpPerspective(image, transform_matrix, (max_width, max_height))
+
+ # return the warped image
+ return warped
+
+ @staticmethod
+ def order_points(pts):
+ rect = np.zeros((4, 2), dtype="float32")
+
+ # the top-left point will have the smallest sum, whereas
+ # the bottom-right point will have the largest sum
+ s = pts.sum(axis=1)
+ rect[0] = pts[np.argmin(s)]
+ rect[2] = pts[np.argmax(s)]
+ diff = np.diff(pts, axis=1)
+ rect[1] = pts[np.argmin(diff)]
+ rect[3] = pts[np.argmax(diff)]
+
+ # return the ordered coordinates
+ return rect
diff --git a/src/utils/interaction.py b/src/utils/interaction.py
new file mode 100644
index 00000000..aba5e754
--- /dev/null
+++ b/src/utils/interaction.py
@@ -0,0 +1,107 @@
+from dataclasses import dataclass
+
+import cv2
+from screeninfo import get_monitors
+
+from src.logger import logger
+from src.utils.image import ImageUtils
+
+monitor_window = get_monitors()[0]
+
+
+@dataclass
+class ImageMetrics:
+ # TODO: Move TEXT_SIZE, etc here and find a better class name
+ window_width, window_height = monitor_window.width, monitor_window.height
+ # for positioning image windows
+ window_x, window_y = 0, 0
+ reset_pos = [0, 0]
+
+
+class InteractionUtils:
+ """Perform primary functions such as displaying images and reading responses"""
+
+ image_metrics = ImageMetrics()
+
+ @staticmethod
+ def show(name, origin, pause=1, resize=False, reset_pos=None, config=None):
+ image_metrics = InteractionUtils.image_metrics
+ if origin is None:
+ logger.info(f"'{name}' - NoneType image to show!")
+ if pause:
+ cv2.destroyAllWindows()
+ return
+ if resize:
+ if not config:
+ raise Exception("config not provided for resizing the image to show")
+ img = ImageUtils.resize_util(origin, config.dimensions.display_width)
+ else:
+ img = origin
+
+ if not is_window_available(name):
+ cv2.namedWindow(name)
+
+ cv2.imshow(name, img)
+
+ if reset_pos:
+ image_metrics.window_x = reset_pos[0]
+ image_metrics.window_y = reset_pos[1]
+
+ cv2.moveWindow(
+ name,
+ image_metrics.window_x,
+ image_metrics.window_y,
+ )
+
+ h, w = img.shape[:2]
+
+ # Set next window position
+ margin = 25
+ w += margin
+ h += margin
+
+ w, h = w // 2, h // 2
+ if image_metrics.window_x + w > image_metrics.window_width:
+ image_metrics.window_x = 0
+ if image_metrics.window_y + h > image_metrics.window_height:
+ image_metrics.window_y = 0
+ else:
+ image_metrics.window_y += h
+ else:
+ image_metrics.window_x += w
+
+ if pause:
+ logger.info(
+ f"Showing '{name}'\n\t Press Q on image to continue. Press Ctrl + C in terminal to exit"
+ )
+
+ wait_q()
+ InteractionUtils.image_metrics.window_x = 0
+ InteractionUtils.image_metrics.window_y = 0
+
+
+@dataclass
+class Stats:
+ # TODO Fill these for stats
+ # Move qbox_vals here?
+ # badThresholds = []
+ # veryBadPoints = []
+ files_moved = 0
+ files_not_moved = 0
+
+
+def wait_q():
+ esc_key = 27
+ while cv2.waitKey(1) & 0xFF not in [ord("q"), esc_key]:
+ pass
+ cv2.destroyAllWindows()
+
+
+def is_window_available(name: str) -> bool:
+ """Checks if a window is available"""
+ try:
+ cv2.getWindowProperty(name, cv2.WND_PROP_VISIBLE)
+ return True
+ except Exception as e:
+ print(e)
+ return False
diff --git a/src/utils/parsing.py b/src/utils/parsing.py
new file mode 100644
index 00000000..cb6901c6
--- /dev/null
+++ b/src/utils/parsing.py
@@ -0,0 +1,113 @@
+import re
+from copy import deepcopy
+from fractions import Fraction
+
+from deepmerge import Merger
+from dotmap import DotMap
+
+from src.constants import FIELD_LABEL_NUMBER_REGEX
+from src.defaults import CONFIG_DEFAULTS, TEMPLATE_DEFAULTS
+from src.schemas.constants import FIELD_STRING_REGEX_GROUPS
+from src.utils.file import load_json
+from src.utils.validations import (
+ validate_config_json,
+ validate_evaluation_json,
+ validate_template_json,
+)
+
+OVERRIDE_MERGER = Merger(
+ # pass in a list of tuples,with the
+ # strategies you are looking to apply
+ # to each type.
+ [
+ # (list, ["prepend"]),
+ (dict, ["merge"])
+ ],
+ # next, choose the fallback strategies,
+ # applied to all other types:
+ ["override"],
+ # finally, choose the strategies in
+ # the case where the types conflict:
+ ["override"],
+)
+
+
+def get_concatenated_response(omr_response, template):
+ # Multi-column/multi-row questions which need to be concatenated
+ concatenated_response = {}
+ for field_label, concatenate_keys in template.custom_labels.items():
+ custom_label = "".join([omr_response[k] for k in concatenate_keys])
+ concatenated_response[field_label] = custom_label
+
+ for field_label in template.non_custom_labels:
+ concatenated_response[field_label] = omr_response[field_label]
+
+ return concatenated_response
+
+
+def open_config_with_defaults(config_path):
+ user_tuning_config = load_json(config_path)
+ user_tuning_config = OVERRIDE_MERGER.merge(
+ deepcopy(CONFIG_DEFAULTS), user_tuning_config
+ )
+ validate_config_json(user_tuning_config, config_path)
+ # https://github.com/drgrib/dotmap/issues/74
+ return DotMap(user_tuning_config, _dynamic=False)
+
+
+def open_template_with_defaults(template_path):
+ user_template = load_json(template_path)
+ user_template = OVERRIDE_MERGER.merge(deepcopy(TEMPLATE_DEFAULTS), user_template)
+ validate_template_json(user_template, template_path)
+ return user_template
+
+
+def open_evaluation_with_validation(evaluation_path):
+ user_evaluation_config = load_json(evaluation_path)
+ validate_evaluation_json(user_evaluation_config, evaluation_path)
+ return user_evaluation_config
+
+
+def parse_fields(key, fields):
+ parsed_fields = []
+ fields_set = set()
+ for field_string in fields:
+ fields_array = parse_field_string(field_string)
+ current_set = set(fields_array)
+ if not fields_set.isdisjoint(current_set):
+ raise Exception(
+ f"Given field string '{field_string}' has overlapping field(s) with other fields in '{key}': {fields}"
+ )
+ fields_set.update(current_set)
+ parsed_fields.extend(fields_array)
+ return parsed_fields
+
+
+def parse_field_string(field_string):
+ if "." in field_string:
+ field_prefix, start, end = re.findall(FIELD_STRING_REGEX_GROUPS, field_string)[
+ 0
+ ]
+ start, end = int(start), int(end)
+ if start >= end:
+ raise Exception(
+ f"Invalid range in fields string: '{field_string}', start: {start} is not less than end: {end}"
+ )
+ return [
+ f"{field_prefix}{field_number}" for field_number in range(start, end + 1)
+ ]
+ else:
+ return [field_string]
+
+
+def custom_sort_output_columns(field_label):
+ label_prefix, label_suffix = re.findall(FIELD_LABEL_NUMBER_REGEX, field_label)[0]
+ return [label_prefix, int(label_suffix) if len(label_suffix) > 0 else 0]
+
+
+def parse_float_or_fraction(result):
+ if type(result) == str and "/" in result:
+ result = float(Fraction(result))
+ else:
+ result = float(result)
+ return result
diff --git a/src/utils/validations.py b/src/utils/validations.py
new file mode 100644
index 00000000..0c2ec839
--- /dev/null
+++ b/src/utils/validations.py
@@ -0,0 +1,115 @@
+"""
+
+ OMRChecker
+
+ Author: Udayraj Deshmukh
+ Github: https://github.com/Udayraj123
+
+"""
+import re
+
+import jsonschema
+from jsonschema import validate
+from rich.table import Table
+
+from src.logger import console, logger
+from src.schemas import SCHEMA_JSONS, SCHEMA_VALIDATORS
+
+
+def validate_evaluation_json(json_data, evaluation_path):
+ logger.info(f"Loading evaluation.json: {evaluation_path}")
+ try:
+ validate(instance=json_data, schema=SCHEMA_JSONS["evaluation"])
+ except jsonschema.exceptions.ValidationError as _err: # NOQA
+ table = Table(show_lines=True)
+ table.add_column("Key", style="cyan", no_wrap=True)
+ table.add_column("Error", style="magenta")
+
+ errors = sorted(
+ SCHEMA_VALIDATORS["evaluation"].iter_errors(json_data),
+ key=lambda e: e.path,
+ )
+ for error in errors:
+ key, validator, msg = parse_validation_error(error)
+ if validator == "required":
+ requiredProperty = re.findall(r"'(.*?)'", msg)[0]
+ table.add_row(
+ f"{key}.{requiredProperty}",
+ msg + ". Make sure the spelling of the key is correct",
+ )
+ else:
+ table.add_row(key, msg)
+ console.print(table, justify="center")
+ raise Exception(
+ f"Provided Evaluation JSON is Invalid: '{evaluation_path}'"
+ ) from None
+
+
+def validate_template_json(json_data, template_path):
+ logger.info(f"Loading template.json: {template_path}")
+ try:
+ validate(instance=json_data, schema=SCHEMA_JSONS["template"])
+ except jsonschema.exceptions.ValidationError as _err: # NOQA
+ table = Table(show_lines=True)
+ table.add_column("Key", style="cyan", no_wrap=True)
+ table.add_column("Error", style="magenta")
+
+ errors = sorted(
+ SCHEMA_VALIDATORS["template"].iter_errors(json_data),
+ key=lambda e: e.path,
+ )
+ for error in errors:
+ key, validator, msg = parse_validation_error(error)
+
+ # Print preProcessor name in case of options error
+ if key == "preProcessors":
+ preProcessorName = json_data["preProcessors"][error.path[1]]["name"]
+ preProcessorKey = error.path[2]
+ table.add_row(f"{key}.{preProcessorName}.{preProcessorKey}", msg)
+ elif validator == "required":
+ requiredProperty = re.findall(r"'(.*?)'", msg)[0]
+ table.add_row(
+ f"{key}.{requiredProperty}",
+ f"{msg}. Check for spelling errors and make sure it is in camelCase",
+ )
+ else:
+ table.add_row(key, msg)
+ console.print(table, justify="center")
+ raise Exception(
+ f"Provided Template JSON is Invalid: '{template_path}'"
+ ) from None
+
+
+def validate_config_json(json_data, config_path):
+ logger.info(f"Loading config.json: {config_path}")
+ try:
+ validate(instance=json_data, schema=SCHEMA_JSONS["config"])
+ except jsonschema.exceptions.ValidationError as _err: # NOQA
+ table = Table(show_lines=True)
+ table.add_column("Key", style="cyan", no_wrap=True)
+ table.add_column("Error", style="magenta")
+ errors = sorted(
+ SCHEMA_VALIDATORS["config"].iter_errors(json_data),
+ key=lambda e: e.path,
+ )
+ for error in errors:
+ key, validator, msg = parse_validation_error(error)
+
+ if validator == "required":
+ requiredProperty = re.findall(r"'(.*?)'", msg)[0]
+ table.add_row(
+ f"{key}.{requiredProperty}",
+ f"{msg}. Check for spelling errors and make sure it is in camelCase",
+ )
+ else:
+ table.add_row(key, msg)
+ console.print(table, justify="center")
+ raise Exception(f"Provided config JSON is Invalid: '{config_path}'") from None
+
+
+def parse_validation_error(error):
+ return (
+ (error.path[0] if len(error.path) > 0 else "$root"),
+ error.validator,
+ error.message,
+ )
diff --git a/template.py b/template.py
deleted file mode 100644
index cdfdb276..00000000
--- a/template.py
+++ /dev/null
@@ -1,355 +0,0 @@
-"""
-
-Designed and Developed by-
-Udayraj Deshmukh
-https://github.com/Udayraj123
-
-"""
-import cv2
-import os
-import json
-import numpy as np
-from globals import *
-from utils import *
-
-### Coordinates Part ###
-
-
-class Pt():
- """
- Container for a Point Box on the OMR
- """
- """
- qNo is the point's property- question to which this point belongs to
- It can be used as a roll number column as well. (eg roll1)
- It can also correspond to a single digit of integer type Q (eg q5d1)
- """
-
- def __init__(self, pt, qNo, qType, val):
- self.x = round(pt[0])
- self.y = round(pt[1])
- self.qNo = qNo
- self.qType = qType
- self.val = val
-
-
-class QBlock():
- def __init__(self, dims, key, orig, traverse_pts):
- # dims = (width, height)
- self.dims = tuple(round(x) for x in dims)
- self.key = key
- self.orig = orig
- self.traverse_pts = traverse_pts
- # will be set when using
- self.shift = 0
-
-
-qtype_data = {
- 'QTYPE_MED': {
- 'vals': ['E', 'H'],
- 'orient': 'V'
- },
- 'QTYPE_ROLL': {
- 'vals': range(10),
- 'orient': 'V'
- },
- 'QTYPE_INT': {
- 'vals': range(10),
- 'orient': 'V'
- },
- 'QTYPE_MCQ4': {
- 'vals': ['A', 'B', 'C', 'D'],
- 'orient': 'H'
- },
- 'QTYPE_MCQ5': {
- 'vals': ['A', 'B', 'C', 'D', 'E'],
- 'orient': 'H'
- },
- # Add custom question types here-
- # ,
- # 'QTYPE_MCQ_COL_5A':{'vals' : ['A']*5, 'orient':'V'},
- # 'QTYPE_MCQ_COL_5B':{'vals' : ['B']*5, 'orient':'V'},
- # 'QTYPE_MCQ_COL_5C':{'vals' : ['C']*5, 'orient':'V'},
- # 'QTYPE_MCQ_COL_5D':{'vals' : ['D']*5, 'orient':'V'},
- # 'QTYPE_MCQ_COL_4A':{'vals' : ['A']*4, 'orient':'V'},
- # 'QTYPE_MCQ_COL_4B':{'vals' : ['B']*4, 'orient':'V'},
- # 'QTYPE_MCQ_COL_4C':{'vals' : ['C']*4, 'orient':'V'},
- # 'QTYPE_MCQ_COL_4D':{'vals' : ['D']*4, 'orient':'V'},
-}
-
-
-class Template():
- def __init__(self, path):
- with open(path, "r") as f:
- json_obj = json.load(f)
- self.path = path
- self.QBlocks = []
- # throw exception on key not exist
- self.dims = json_obj["Dimensions"]
- self.bubbleDims = json_obj["BubbleDimensions"]
- self.concats = json_obj["Concatenations"]
- self.singles = json_obj["Singles"]
-
- # Add new qTypes from template
- if "qTypes" in json_obj:
- qtype_data.update(json_obj["qTypes"])
-
- # process local options
- self.options = json_obj.get("Options", {})
-
- self.marker = None
- self.marker_path = None
- # process markers
- if "Marker" in self.options:
- markerOps = self.options["Marker"]
- self.marker_path = os.path.join(
- os.path.dirname(path), markerOps.get(
- "RelativePath", MARKER_FILE))
- if(not os.path.exists(self.marker_path)):
- print(
- "Error: Marker not found at path provided in template:",
- self.marker_path)
- exit(31)
-
- marker = cv2.imread(self.marker_path, cv2.IMREAD_GRAYSCALE)
- if("SheetToMarkerWidthRatio" in markerOps):
- marker = resize_util(marker, uniform_width /
- int(markerOps["SheetToMarkerWidthRatio"]))
- marker = cv2.GaussianBlur(marker, (5, 5), 0)
- marker = cv2.normalize(
- marker,
- None,
- alpha=0,
- beta=255,
- norm_type=cv2.NORM_MINMAX)
- # marker_eroded_sub = marker-cv2.erode(marker,None)
- self.marker = marker - \
- cv2.erode(marker, kernel=np.ones((5, 5)), iterations=5)
-
- # Allow template to override globals
- # TODO: This is a hack as there should no be any global configuration
- # All template configuration should be local. Global config should
- # be via command args.
- self.globals = json_obj.get("Globals")
- self.update_globals()
-
- # Add QBlocks
- for name, block in json_obj["QBlocks"].items():
- self.addQBlocks(name, block)
-
- def update_globals(self):
- if self.globals:
- globals().update(self.globals)
-
- # Expects bubbleDims to be set already
- def addQBlocks(self, key, rect):
- assert(self.bubbleDims != [-1, -1])
- # For qType defined in QBlocks
- if 'qType' in rect:
- rect.update(**qtype_data[rect['qType']])
- else:
- rect['qType'] = {'vals': rect['vals'], 'orient': rect['orient']}
- # keyword arg unpacking followed by named args
- self.QBlocks += genGrid(self.bubbleDims, key, **rect)
- # self.QBlocks.append(QBlock(rect.orig, calcQBlockDims(rect), maketemplate(rect)))
-
-
-def genQBlock(
- bubbleDims,
- QBlockDims,
- key,
- orig,
- qNos,
- gaps,
- vals,
- qType,
- orient,
- col_orient):
- """
- Input:
- orig - start point
- qNos - a tuple of qNos
- gaps - (gapX,gapY) are the gaps between rows and cols in a block
- vals - a 1D array of values of each alternative for a question
-
- Output:
- // Returns set of coordinates of a rectangular grid of points
- Returns a QBlock containing array of Qs and some metadata?!
-
- Ref:
- 1 2 3 4
- 1 2 3 4
- 1 2 3 4
-
- (q1, q2, q3)
-
- 00
- 11
- 22
- 33
- 44
-
- (q1.1,q1.2)
-
- """
- H, V = (0, 1) if(orient == 'H') else (1, 0)
- # orig[0] += np.random.randint(-6,6)*2 # test random shift
- traverse_pts = []
- o = [float(i) for i in orig]
-
- if(col_orient == orient):
- for q in range(len(qNos)):
- pt = o.copy()
- pts = []
- for v in range(len(vals)):
- pts.append(Pt(pt.copy(), qNos[q], qType, vals[v]))
- pt[H] += gaps[H]
- # For diagonalal endpoint of QBlock
- pt[H] += bubbleDims[H] - gaps[H]
- pt[V] += bubbleDims[V]
- # TODO- make a mini object for this
- traverse_pts.append(([o.copy(), pt.copy()], pts))
- o[V] += gaps[V]
- else:
- for v in range(len(vals)):
- pt = o.copy()
- pts = []
- for q in range(len(qNos)):
- pts.append(Pt(pt.copy(), qNos[q], qType, vals[v]))
- pt[V] += gaps[V]
- # For diagonalal endpoint of QBlock
- pt[V] += bubbleDims[V] - gaps[V]
- pt[H] += bubbleDims[H]
- # TODO- make a mini object for this
- traverse_pts.append(([o.copy(), pt.copy()], pts))
- o[H] += gaps[H]
- # Pass first three args as is. only append 'traverse_pts'
- return QBlock(QBlockDims, key, orig, traverse_pts)
-
-
-def genGrid(
- bubbleDims,
- key,
- qType,
- orig,
- bigGaps,
- gaps,
- qNos,
- vals,
- orient='V',
- col_orient='V'):
- """
- Input(Directly passable from JSON parameters):
- bubbleDims - dimesions of single QBox
- orig- start point
- qNos - an array of qNos tuples(see below) that align with dimension of the big grid (gridDims extracted from here)
- bigGaps - (bigGapX,bigGapY) are the gaps between blocks
- gaps - (gapX,gapY) are the gaps between rows and cols in a block
- vals - a 1D array of values of each alternative for a question
- orient - The way of arranging the vals (vertical or horizontal)
-
- Output:
- // Returns an array of Q objects (having their points) arranged in a rectangular grid
- Returns grid of QBlock objects
-
- 00 00 00 00
- Q1 1 2 3 4 1 2 3 4 11 11 11 11
- Q2 1 2 3 4 1 2 3 4 22 22 22 22 1234567
- Q3 1 2 3 4 1 2 3 4 33 33 33 33 1234567
- 44 44 44 44
- , 55 55 55 55 , 1234567 and many more possibilities!
- Q7 1 2 3 4 1 2 3 4 66 66 66 66 1234567
- Q8 1 2 3 4 1 2 3 4 77 77 77 77
- Q9 1 2 3 4 1 2 3 4 88 88 88 88
- 99 99 99 99
-
-TODO: Update this part, add more examples like-
- Q1 1 2 3 4
-
- Q2 1 2 3 4
- Q3 1 2 3 4
-
- Q4 1 2 3 4
- Q5 1 2 3 4
-
- MCQ type (orient='H')-
- [
- [(q1,q2,q3),(q4,q5,q6)]
- [(q7,q8,q9),(q10,q11,q12)]
- ]
-
- INT type (orient='V')-
- [
- [(q1d1,q1d2),(q2d1,q2d2),(q3d1,q3d2),(q4d1,q4d2)]
- ]
-
- ROLL type-
- [
- [(roll1,roll2,roll3,...,roll10)]
- ]
-
- """
- gridData = np.array(qNos)
- # print(gridData.shape, gridData)
- if(0 and len(gridData.shape) != 3 or gridData.size == 0): # product of shape is zero
- print(
- "Error(genGrid): Invalid qNos array given:",
- gridData.shape,
- gridData)
- exit(32)
-
- orig = np.array(orig)
- numQsMax = max([max([len(qb) for qb in row]) for row in gridData])
-
- numDims = [numQsMax, len(vals)]
-
- QBlocks = []
-
- # **Simple is powerful**
- # H and V are named with respect to orient == 'H', reverse their meaning
- # when orient = 'V'
- H, V = (0, 1) if(orient == 'H') else (1, 0)
-
- # print(orig, numDims, gridData.shape, gridData)
- # orient is also the direction of making QBlocks
-
- # print(key, numDims, orig, gaps, bigGaps, origGap )
- qStart = orig.copy()
-
- origGap = [0, 0]
-
- # Usually single row
- for row in gridData:
- qStart[V] = orig[V]
-
- # Usually multiple qTuples
- for qTuple in row:
- # Update numDims and origGaps
- numDims[0] = len(qTuple)
- # bigGaps is indep of orientation
- origGap[0] = bigGaps[0] + (numDims[V] - 1) * gaps[H]
- origGap[1] = bigGaps[1] + (numDims[H] - 1) * gaps[V]
- # each qTuple will have qNos
- QBlockDims = [
- # width x height in pixels
- gaps[0] * (numDims[V] - 1) + bubbleDims[H],
- gaps[1] * (numDims[H] - 1) + bubbleDims[V]
- ]
- # WATCH FOR BLUNDER(use .copy()) - qStart was getting passed by
- # reference! (others args read-only)
- QBlocks.append(
- genQBlock(
- bubbleDims,
- QBlockDims,
- key,
- qStart.copy(),
- qTuple,
- gaps,
- vals,
- qType,
- orient,
- col_orient))
- # Goes vertically down first
- qStart[V] += origGap[V]
- qStart[H] += origGap[H]
- return QBlocks
diff --git a/utils.py b/utils.py
deleted file mode 100644
index 25f6df64..00000000
--- a/utils.py
+++ /dev/null
@@ -1,1168 +0,0 @@
-"""
-
-Designed and Developed by-
-Udayraj Deshmukh
-https://github.com/Udayraj123
-
-"""
-from time import localtime, strftime, time
-from random import randint
-from template import *
-from globals import *
-
-# In[62]:
-import re
-import os
-import sys
-import cv2
-import glob
-from imutils import grab_contours
-import numpy as np
-import pandas as pd
-import matplotlib.pyplot as plt
-plt.rcParams['figure.figsize'] = (10.0, 8.0)
-
-
-def setup_dirs(paths):
- print('\nChecking Directories...')
- for _dir in [paths.saveMarkedDir]:
- if(not os.path.exists(_dir)):
- print('Created : ' + _dir)
- os.makedirs(_dir)
- os.mkdir(_dir + '/stack')
- os.mkdir(_dir + '/_MULTI_')
- os.mkdir(_dir + '/_MULTI_' + '/stack')
- # os.mkdir(_dir+sl+'/_BADSCAN_')
- # os.mkdir(_dir+sl+'/_BADSCAN_'+'/stack')
- else:
- print('Present : ' + _dir)
-
- for _dir in [paths.manualDir, paths.resultDir]:
- if(not os.path.exists(_dir)):
- print('Created : ' + _dir)
- os.makedirs(_dir)
- else:
- print('Present : ' + _dir)
-
- for _dir in [paths.multiMarkedDir, paths.errorsDir, paths.badRollsDir]:
- if(not os.path.exists(_dir)):
- print('Created : ' + _dir)
- os.makedirs(_dir)
- else:
- print('Present : ' + _dir)
-
-
-def waitQ():
- ESC_KEY = 27
- while(cv2.waitKey(1) & 0xFF not in [ord('q'), ESC_KEY]):
- pass
- cv2.destroyAllWindows()
-
-
-def normalize_util(img, alpha=0, beta=255):
- return cv2.normalize(img, alpha, beta, norm_type=cv2.NORM_MINMAX)
-
-
-def normalize_hist(img):
- hist, bins = np.histogram(img.flatten(), 256, [0, 256])
- cdf = hist.cumsum()
- cdf_m = np.ma.masked_equal(cdf, 0)
- cdf_m = (cdf_m - cdf_m.min()) * 255 / (cdf_m.max() - cdf_m.min())
- cdf = np.ma.filled(cdf_m, 0).astype('uint8')
- return cdf[img]
-
-
-def resize_util(img, u_width, u_height=None):
- if u_height is None:
- h, w = img.shape[:2]
- u_height = int(h * u_width / w)
- return cv2.resize(img, (int(u_width), int(u_height)))
-
-
-def resize_util_h(img, u_height, u_width=None):
- if u_width is None:
- h, w = img.shape[:2]
- u_width = int(w * u_height / h)
- return cv2.resize(img, (int(u_width), int(u_height)))
-
-
-def show(name, orig, pause=1, resize=False, resetpos=None):
- global windowX, windowY, display_width
- if(type(orig) == type(None)):
- print(name, " NoneType image to show!")
- if(pause):
- cv2.destroyAllWindows()
- return
- origDim = orig.shape[:2]
- img = resize_util(orig, display_width, display_height) if resize else orig
- cv2.imshow(name, img)
- if(resetpos):
- windowX = resetpos[0]
- windowY = resetpos[1]
- cv2.moveWindow(name, windowX, windowY)
-
- h, w = img.shape[:2]
-
- # Set next window position
- margin = 25
- w += margin
- h += margin
- if(windowX + w > windowWidth):
- windowX = 0
- if(windowY + h > windowHeight):
- windowY = 0
- else:
- windowY += h
- else:
- windowX += w
-
- if(pause):
- print(
- "Showing '" +
- name +
- "'\n\tPress Q on image to continue Press Ctrl + C in terminal to exit")
- waitQ()
-
-
-def putLabel(img, label, size):
- scale = img.shape[1] / display_width
- bgVal = int(np.mean(img))
- pos = (int(scale * 80), int(scale * 30))
- clr = (255 - bgVal,) * 3
- img[(pos[1] - size * 30):(pos[1] + size * 2), :] = bgVal
- cv2.putText(img, label, pos, cv2.FONT_HERSHEY_SIMPLEX, size, clr, 3)
-
-
-def drawTemplateLayout(
- img,
- template,
- shifted=True,
- draw_qvals=False,
- border=-1):
- img = resize_util(img, template.dims[0], template.dims[1])
- final_align = img.copy()
- boxW, boxH = template.bubbleDims
- for QBlock in template.QBlocks:
- s, d = QBlock.orig, QBlock.dims
- shift = QBlock.shift
- if(shifted):
- cv2.rectangle(
- final_align,
- (s[0] + shift,
- s[1]),
- (s[0] + shift + d[0],
- s[1] + d[1]),
- CLR_BLACK,
- 3)
- else:
- cv2.rectangle(
- final_align, (s[0], s[1]), (s[0] + d[0], s[1] + d[1]), CLR_BLACK, 3)
- for qStrip, qBoxPts in QBlock.traverse_pts:
- for pt in qBoxPts:
- x, y = (pt.x + QBlock.shift, pt.y) if shifted else (pt.x, pt.y)
- cv2.rectangle(final_align,
- (int(x + boxW / 10),
- int(y + boxH / 10)),
- (int(x + boxW - boxW / 10),
- int(y + boxH - boxH / 10)),
- CLR_GRAY,
- border)
- if(draw_qvals):
- rect = [y, y + boxH, x, x + boxW]
- cv2.putText(final_align, '%d' % (cv2.mean(img[rect[0]:rect[1], rect[2]:rect[3]])[
- 0]), (rect[2] + 2, rect[0] + (boxH * 2) // 3), cv2.FONT_HERSHEY_SIMPLEX, 0.6, CLR_BLACK, 2)
- if(shifted):
- cv2.putText(final_align, 's%s' %
- (shift), tuple(s -
- [template.dims[0] //
- 20, -
- d[1] //
- 2]), cv2.FONT_HERSHEY_SIMPLEX, TEXT_SIZE, CLR_BLACK, 4)
- return final_align
-
-
-def getPlotImg():
- plt.savefig('tmp.png')
- # img = cv2.imread('tmp.png',cv2.IMREAD_COLOR)
- img = cv2.imread('tmp.png', cv2.IMREAD_GRAYSCALE)
- os.remove("tmp.png")
- # plt.cla()
- # plt.clf()
- plt.close()
- return img
-
-
-def order_points(pts):
- rect = np.zeros((4, 2), dtype="float32")
-
- # the top-left point will have the smallest sum, whereas
- # the bottom-right point will have the largest sum
- s = pts.sum(axis=1)
- rect[0] = pts[np.argmin(s)]
- rect[2] = pts[np.argmax(s)]
- diff = np.diff(pts, axis=1)
- rect[1] = pts[np.argmin(diff)]
- rect[3] = pts[np.argmax(diff)]
-
- # return the ordered coordinates
- return rect
-
-
-def four_point_transform(image, pts):
- # obtain a consistent order of the points and unpack them
- # individually
- rect = order_points(pts)
- (tl, tr, br, bl) = rect
-
- # compute the width of the new image, which will be the
- widthA = np.sqrt(((br[0] - bl[0]) ** 2) + ((br[1] - bl[1]) ** 2))
- widthB = np.sqrt(((tr[0] - tl[0]) ** 2) + ((tr[1] - tl[1]) ** 2))
-
- maxWidth = max(int(widthA), int(widthB))
- # maxWidth = max(int(np.linalg.norm(br-bl)), int(np.linalg.norm(tr-tl)))
-
- # compute the height of the new image, which will be the
- heightA = np.sqrt(((tr[0] - br[0]) ** 2) + ((tr[1] - br[1]) ** 2))
- heightB = np.sqrt(((tl[0] - bl[0]) ** 2) + ((tl[1] - bl[1]) ** 2))
- maxHeight = max(int(heightA), int(heightB))
- # maxHeight = max(int(np.linalg.norm(tr-br)), int(np.linalg.norm(tl-br)))
-
- # now that we have the dimensions of the new image, construct
- # the set of destination points to obtain a "birds eye view",
- # (i.e. top-down view) of the image, again specifying points
- # in the top-left, top-right, bottom-right, and bottom-left
- # order
- dst = np.array([
- [0, 0],
- [maxWidth - 1, 0],
- [maxWidth - 1, maxHeight - 1],
- [0, maxHeight - 1]], dtype="float32")
-
- # compute the perspective transform matrix and then apply it
- M = cv2.getPerspectiveTransform(rect, dst)
- warped = cv2.warpPerspective(image, M, (maxWidth, maxHeight))
-
- # return the warped image
- return warped
-
-
-def dist(p1, p2):
- return np.linalg.norm(np.array(p1) - np.array(p2))
-
-
-def get_reflection(pt, pt1, pt2):
- pt, pt1, pt2 = tuple(
- map(lambda x: np.array(x, dtype=float), [pt, pt1, pt2]))
- return (pt1 + pt2) - pt
-
-
-def printbuf(x):
- sys.stdout.write(str(x))
- sys.stdout.write('\r')
-
-
-def get_fourth_pt(three_pts):
- m = []
- for i in range(3):
- m.append(dist(three_pts[i], three_pts[(i + 1) % 3]))
-
- v = max(m)
- for i in range(3):
- if(m[i] != v and m[(i + 1) % 3] != v):
- refl = (i + 1) % 3
- break
- fourth_pt = get_reflection(
- three_pts[refl], three_pts[(refl + 1) % 3], three_pts[(refl + 2) % 3])
- return fourth_pt
-
-
-def angle(p1, p2, p0):
- dx1 = float(p1[0] - p0[0])
- dy1 = float(p1[1] - p0[1])
- dx2 = float(p2[0] - p0[0])
- dy2 = float(p2[1] - p0[1])
- return (dx1 * dx2 + dy1 * dy2) / \
- np.sqrt((dx1 * dx1 + dy1 * dy1) * (dx2 * dx2 + dy2 * dy2) + 1e-10)
-
-
-def checkMaxCosine(approx):
- # assumes 4 pts present
- maxCosine = 0
- minCosine = 1.5
- for i in range(2, 5):
- cosine = abs(angle(approx[i % 4], approx[i - 2], approx[i - 1]))
- maxCosine = max(cosine, maxCosine)
- minCosine = min(cosine, minCosine)
- # TODO add to plot dict
- # print(maxCosine)
- if(maxCosine >= 0.35):
- print('Quadrilateral is not a rectangle.')
- return False
- return True
-
-
-def validateRect(approx):
- # TODO: add logic from app?!
- return len(approx) == 4 and checkMaxCosine(approx.reshape(4, 2))
-
-
-def auto_canny(image, sigma=0.93):
- # compute the median of the single channel pixel intensities
- v = np.median(image)
-
- # apply automatic Canny edge detection using the computed median
- lower = int(max(0, (1.0 - sigma) * v))
- upper = int(min(255, (1.0 + sigma) * v))
- edged = cv2.Canny(image, lower, upper)
-
- # return the edged image
- return edged
-
-
-def resetSaveImg(key):
- global saveImgList
- saveImgList[key] = []
-
-
-def appendSaveImg(key, img):
- if(saveimglvl >= int(key)):
- global saveImgList
- if(key not in saveImgList):
- saveImgList[key] = []
- saveImgList[key].append(img.copy())
-
-
-def findPage(image_norm):
- # Done: find ORIGIN for the quadrants
- # Done, Auto tune! : Get canny parameters tuned
- # (https://www.pyimagesearch.com/2015/04/06/zero-parameter-automatic-canny-edge-detection-with-python-and-opencv/)
-
- image_norm = normalize_util(image_norm)
- ret, image_norm = cv2.threshold(image_norm, 200, 255, cv2.THRESH_TRUNC)
- image_norm = normalize_util(image_norm)
-
- appendSaveImg(1, image_norm)
-
- # kernel = cv2.getStructuringElement(cv2.MORPH_RECT, (3, 10))
- kernel = cv2.getStructuringElement(cv2.MORPH_RECT, (10, 10))
- """
- # Closing is reverse of Opening, Dilation followed by Erosion.
- A pixel in the original image (either 1 or 0) will be considered 1 only if all the pixels
- under the kernel is 1, otherwise it is eroded (made to zero).
- """
- # Close the small holes, i.e. Complete the edges on canny image
- closed = cv2.morphologyEx(image_norm, cv2.MORPH_CLOSE, kernel)
-
- appendSaveImg(1, closed)
-
- edge = cv2.Canny(closed, 185, 55)
-
- # findContours returns outer boundaries in CW and inner boundaries in ACW
- # order.
- cnts = grab_contours(
- cv2.findContours(
- edge,
- cv2.RETR_LIST,
- cv2.CHAIN_APPROX_SIMPLE))
- # hullify to resolve disordered curves due to noise
- cnts = [cv2.convexHull(c) for c in cnts]
- cnts = sorted(cnts, key=cv2.contourArea, reverse=True)[:5]
- sheet = []
- for c in cnts:
- if cv2.contourArea(c) < MIN_PAGE_AREA:
- continue
- peri = cv2.arcLength(c, True)
- # ez algo -
- # https://en.wikipedia.org/wiki/Ramer–Douglas–Peucker_algorithm
- approx = cv2.approxPolyDP(c, epsilon=0.025 * peri, closed=True)
- # print("Area",cv2.contourArea(c), "Peri", peri)
-
- # check its rectangle-ness:
- if(validateRect(approx)):
- sheet = np.reshape(approx, (4, -1))
- cv2.drawContours(image_norm, [approx], -1, (0, 255, 0), 2)
- cv2.drawContours(edge, [approx], -1, (255, 255, 255), 10)
- break
- # box = perspective.order_points(box)
- # sobel = cv2.addWeighted(cv2.Sobel(edge, cv2.CV_64F, 1, 0, ksize=3),0.5,cv2.Sobel(edge, cv2.CV_64F, 0, 1, ksize=3),0.5,0,edge)
- # ExcessDo : make it work on killer images
- # edge2 = auto_canny(image_norm)
- # show('Morphed Edges',np.hstack((closed,edge)),1,1)
-
- appendSaveImg(1, edge)
- return sheet
-
-
-# Resizing the marker within scaleRange at rate of descent_per_step to
-# find the best match.
-def getBestMatch(image_eroded_sub, marker):
-
- descent_per_step = (
- marker_rescale_range[1] - marker_rescale_range[0]) // marker_rescale_steps
- h, w = marker.shape[:2]
- res, best_scale = None, None
- allMaxT = 0
-
- for r0 in np.arange(
- marker_rescale_range[1], marker_rescale_range[0], -1 * descent_per_step): # reverse order
- s = float(r0 * 1 / 100)
- if(s == 0.0):
- continue
- rescaled_marker = resize_util_h(
- marker if ERODE_SUB_OFF else marker,
- u_height=int(
- h * s))
- # res is the black image with white dots
- res = cv2.matchTemplate(
- image_eroded_sub,
- rescaled_marker,
- cv2.TM_CCOEFF_NORMED)
-
- maxT = res.max()
- if(allMaxT < maxT):
- # print('Scale: '+str(s)+', Circle Match: '+str(round(maxT*100,2))+'%')
- best_scale, allMaxT = s, maxT
-
- if(allMaxT < thresholdCircle):
- print("\tWarning: Template matching too low! Should you pass --noCropping flag?")
- if(showimglvl >= 1):
- show("res", res, 1, 0)
-
- if(best_scale is None):
- print("No matchings for given scaleRange:", marker_rescale_range)
- return best_scale, allMaxT
-
-
-def adjust_gamma(image, gamma=1.0):
- # build a lookup table mapping the pixel values [0, 255] to
- # their adjusted gamma values
- invGamma = 1.0 / gamma
- table = np.array([((i / 255.0) ** invGamma) * 255
- for i in np.arange(0, 256)]).astype("uint8")
-
- # apply gamma correction using the lookup table
- return cv2.LUT(image, table)
-
-
-clahe = cv2.createCLAHE(clipLimit=5.0, tileGridSize=(8, 8))
-# TODO Fill these for stats
-thresholdCircles = []
-badThresholds = []
-veryBadPoints = []
-
-
-def getROI(image, filename, noCropping=False):
- global clahe
- for i in range(saveimglvl):
- resetSaveImg(i + 1)
-
- appendSaveImg(1, image)
- """
- TODO later Autorotate:
- - Rotate 90 : check page width:height, CW/ACW? - do CW, then pass to 180 check.
- - Rotate 180 :
- Nope, OMR specific, paper warping may be imperfect. - check markers centroid
- Nope - OCR check
- Match logo - can work, but 'lon' too big and may unnecessarily rotate? - but you know the scale
- Check roll field morphed
- """
-
- # TODO: (remove noCropping bool) Automate the case of close up scan(incorrect page)-
- # ^Note: App rejects croppeds along with others
-
- # image = resize_util(image, uniform_width, uniform_height)
-
- # Preprocessing the image
- img = image.copy()
- # TODO: need to detect if image is too blurry already! (M1: check
- # noCropping dimensions b4 resizing coz it won't be blurry otherwise _/)
- img = cv2.GaussianBlur(img, (3, 3), 0)
- image_norm = normalize_util(img)
-
- if(noCropping == False):
- # Need this resize for arbitrary high res images: before passing to
- # findPage
- if(image_norm.shape[1] > uniform_width * 2):
- image_norm = resize_util(image_norm, uniform_width * 2)
- sheet = findPage(image_norm)
- if sheet == []:
- print("\tError: Paper boundary not found! Should you pass --noCropping flag?")
- return None
- else:
- print("Found page corners: \t", sheet.tolist())
-
- # Warp layer 1
- image_norm = four_point_transform(image_norm, sheet)
-
- # Resize only after cropping the page for clarity as well as uniformity
- # for non noCropping images
- image_norm = resize_util(image_norm, uniform_width, uniform_height)
- image = resize_util(image, uniform_width, uniform_height)
- appendSaveImg(1, image_norm)
-
- # Return preprocessed image
- return image_norm
-
-
-def handle_markers(image_norm, marker, curr_filename):
-
- if ERODE_SUB_OFF:
- image_eroded_sub = normalize_util(image_norm)
- else:
- image_eroded_sub = normalize_util(image_norm
- - cv2.erode(image_norm,
- kernel=np.ones((5, 5)),
- iterations=5))
- # Quads on warped image
- quads = {}
- h1, w1 = image_eroded_sub.shape[:2]
- midh, midw = h1 // 3, w1 // 2
- origins = [[0, 0], [midw, 0], [0, midh], [midw, midh]]
- quads[0] = image_eroded_sub[0:midh, 0:midw]
- quads[1] = image_eroded_sub[0:midh, midw:w1]
- quads[2] = image_eroded_sub[midh:h1, 0:midw]
- quads[3] = image_eroded_sub[midh:h1, midw:w1]
-
- # Draw Quadlines
- image_eroded_sub[:, midw:midw + 2] = 255
- image_eroded_sub[midh:midh + 2, :] = 255
-
- best_scale, allMaxT = getBestMatch(image_eroded_sub, marker)
- if(best_scale is None):
- # TODO: Plot and see performance of marker_rescale_range
- if(showimglvl >= 1):
- show('Quads', image_eroded_sub)
- return None
-
- optimal_marker = resize_util_h(
- marker if ERODE_SUB_OFF else marker, u_height=int(
- marker.shape[0] * best_scale))
- h, w = optimal_marker.shape[:2]
- centres = []
- sumT, maxT = 0, 0
- print("Matching Marker:\t", end=" ")
- for k in range(0, 4):
- res = cv2.matchTemplate(quads[k], optimal_marker, cv2.TM_CCOEFF_NORMED)
- maxT = res.max()
- print("Q" + str(k + 1) + ": maxT", round(maxT, 3), end="\t")
- if(maxT < thresholdCircle or abs(allMaxT - maxT) >= thresholdVar):
- # Warning - code will stop in the middle. Keep Threshold low to
- # avoid.
- print(
- curr_filename,
- "\nError: No circle found in Quad",
- k + 1,
- "\n\tthresholdVar",
- thresholdVar,
- "maxT",
- maxT,
- "allMaxT",
- allMaxT,
- "Should you pass --noCropping flag?")
- if(showimglvl >= 1):
- show("no_pts_" + curr_filename, image_eroded_sub, 0)
- show("res_Q" + str(k + 1), res, 1)
- return None
-
- pt = np.argwhere(res == maxT)[0]
- pt = [pt[1], pt[0]]
- pt[0] += origins[k][0]
- pt[1] += origins[k][1]
- # print(">>",pt)
- image_norm = cv2.rectangle(image_norm, tuple(
- pt), (pt[0] + w, pt[1] + h), (150, 150, 150), 2)
- # display:
- image_eroded_sub = cv2.rectangle(
- image_eroded_sub,
- tuple(pt),
- (pt[0] + w,
- pt[1] + h),
- (50,
- 50,
- 50) if ERODE_SUB_OFF else (
- 155,
- 155,
- 155),
- 4)
- centres.append([pt[0] + w / 2, pt[1] + h / 2])
- sumT += maxT
- print("Optimal Scale:", best_scale)
- # analysis data
- thresholdCircles.append(sumT / 4)
-
- image_norm = four_point_transform(image_norm, np.array(centres))
- # appendSaveImg(1,image_eroded_sub)
- # appendSaveImg(1,image_norm)
-
- appendSaveImg(2, image_eroded_sub)
- # Debugging image -
- # res = cv2.matchTemplate(image_eroded_sub,optimal_marker,cv2.TM_CCOEFF_NORMED)
- # res[ : , midw:midw+2] = 255
- # res[ midh:midh+2, : ] = 255
- # show("Markers Matching",res)
- if(showimglvl >= 2 and showimglvl < 4):
- image_eroded_sub = resize_util_h(image_eroded_sub, image_norm.shape[0])
- image_eroded_sub[:, -5:] = 0
- h_stack = np.hstack((image_eroded_sub, image_norm))
- show("Warped: " + curr_filename, resize_util(h_stack,
- int(display_width * 1.6)), 0, 0, [0, 0])
- # iterations : Tuned to 2.
- # image_eroded_sub = image_norm - cv2.erode(image_norm, kernel=np.ones((5,5)),iterations=2)
- return image_norm
-
-
-def getGlobalThreshold(
- QVals_orig,
- plotTitle=None,
- plotShow=True,
- sortInPlot=True,
- looseness=1):
- """
- Note: Cannot assume qStrip has only-gray or only-white bg (in which case there is only one jump).
- So there will be either 1 or 2 jumps.
- 1 Jump :
- ......
- ||||||
- |||||| <-- risky THR
- |||||| <-- safe THR
- ....||||||
- ||||||||||
-
- 2 Jumps :
- ......
- |||||| <-- wrong THR
- ....||||||
- |||||||||| <-- safe THR
- ..||||||||||
- ||||||||||||
-
- The abstract "First LARGE GAP" is perfect for this.
- Current code is considering ONLY TOP 2 jumps(>= MIN_GAP) to be big, gives the smaller one
-
- """
- # Sort the Q vals
- QVals = sorted(QVals_orig)
- # Find the FIRST LARGE GAP and set it as threshold:
- ls = (looseness + 1) // 2
- l = len(QVals) - ls
- max1, thr1 = MIN_JUMP, 255
- for i in range(ls, l):
- jump = QVals[i + ls] - QVals[i - ls]
- if(jump > max1):
- max1 = jump
- thr1 = QVals[i - ls] + jump / 2
-
-# NOTE: thr2 is deprecated, thus is JUMP_DELTA
- # Make use of the fact that the JUMP_DELTA(Vertical gap ofc) between
- # values at detected jumps would be atleast 20
- max2, thr2 = MIN_JUMP, 255
- # Requires atleast 1 gray box to be present (Roll field will ensure this)
- for i in range(ls, l):
- jump = QVals[i + ls] - QVals[i - ls]
- newThr = QVals[i - ls] + jump / 2
- if(jump > max2 and abs(thr1 - newThr) > JUMP_DELTA):
- max2 = jump
- thr2 = newThr
- # globalTHR = min(thr1,thr2)
- globalTHR, j_low, j_high = thr1, thr1 - max1 // 2, thr1 + max1 // 2
-
- # # For normal images
- # thresholdRead = 116
- # if(thr1 > thr2 and thr2 > thresholdRead):
- # print("Note: taking safer thr line.")
- # globalTHR, j_low, j_high = thr2, thr2 - max2//2, thr2 + max2//2
-
- if(plotTitle is not None):
- f, ax = plt.subplots()
- ax.bar(range(len(QVals_orig)), QVals if sortInPlot else QVals_orig)
- ax.set_title(plotTitle)
- thrline = ax.axhline(globalTHR, color='green', ls='--', linewidth=5)
- thrline.set_label("Global Threshold")
- thrline = ax.axhline(thr2, color='red', ls=':', linewidth=3)
- thrline.set_label("THR2 Line")
- # thrline=ax.axhline(j_low,color='red',ls='-.', linewidth=3)
- # thrline=ax.axhline(j_high,color='red',ls='-.', linewidth=3)
- # thrline.set_label("Boundary Line")
- # ax.set_ylabel("Mean Intensity")
- ax.set_ylabel("Values")
- ax.set_xlabel("Position")
- ax.legend()
- if(plotShow):
- plt.title(plotTitle)
- plt.show()
-
- return globalTHR, j_low, j_high
-
-
-def getLocalThreshold(
- qNo,
- QVals,
- globalTHR,
- noOutliers,
- plotTitle=None,
- plotShow=True):
- """
- TODO: Update this documentation too-
- //No more - Assumption : Colwise background color is uniformly gray or white, but not alternating. In this case there is atmost one jump.
-
- 0 Jump :
- <-- safe THR?
- .......
- ...|||||||
- |||||||||| <-- safe THR?
- // How to decide given range is above or below gray?
- -> global QVals shall absolutely help here. Just run same function on total QVals instead of colwise _//
- How to decide it is this case of 0 jumps
-
- 1 Jump :
- ......
- ||||||
- |||||| <-- risky THR
- |||||| <-- safe THR
- ....||||||
- ||||||||||
-
- """
- # Sort the Q vals
- QVals = sorted(QVals)
-
- # Small no of pts cases:
- # base case: 1 or 2 pts
- if(len(QVals) < 3):
- thr1 = globalTHR if np.max(
- QVals) - np.min(QVals) < MIN_GAP else np.mean(QVals)
- else:
- # qmin, qmax, qmean, qstd = round(np.min(QVals),2), round(np.max(QVals),2), round(np.mean(QVals),2), round(np.std(QVals),2)
- # GVals = [round(abs(q-qmean),2) for q in QVals]
- # gmean, gstd = round(np.mean(GVals),2), round(np.std(GVals),2)
- # # DISCRETION: Pretty critical factor in reading response
- # # Doesn't work well for small number of values.
- # DISCRETION = 2.7 # 2.59 was closest hit, 3.0 is too far
- # L2MaxGap = round(max([abs(g-gmean) for g in GVals]),2)
- # if(L2MaxGap > DISCRETION*gstd):
- # noOutliers = False
-
- # # ^Stackoverflow method
- # print(qNo, noOutliers,"qstd",round(np.std(QVals),2), "gstd", gstd,"Gaps in gvals",sorted([round(abs(g-gmean),2) for g in GVals],reverse=True), '\t',round(DISCRETION*gstd,2), L2MaxGap)
-
- # else:
- # Find the LARGEST GAP and set it as threshold: //(FIRST LARGE GAP)
- l = len(QVals) - 1
- max1, thr1 = MIN_JUMP, 255
- for i in range(1, l):
- jump = QVals[i + 1] - QVals[i - 1]
- if(jump > max1):
- max1 = jump
- thr1 = QVals[i - 1] + jump / 2
- # print(qNo,QVals,max1)
-
- # If not confident, then only take help of globalTHR
- if(max1 < CONFIDENT_JUMP):
- if(noOutliers):
- # All Black or All White case
- thr1 = globalTHR
- else:
- # TODO: Low confidence parameters here
- pass
-
- # if(thr1 == 255):
- # print("Warning: threshold is unexpectedly 255! (Outlier Delta issue?)",plotTitle)
-
- if(plotShow and plotTitle is not None):
- f, ax = plt.subplots()
- ax.bar(range(len(QVals)), QVals)
- thrline = ax.axhline(thr1, color='green', ls=('-.'), linewidth=3)
- thrline.set_label("Local Threshold")
- thrline = ax.axhline(globalTHR, color='red', ls=':', linewidth=5)
- thrline.set_label("Global Threshold")
- ax.set_title(plotTitle)
- ax.set_ylabel("Bubble Mean Intensity")
- ax.set_xlabel("Bubble Number(sorted)")
- ax.legend()
- # TODO append QStrip to this plot-
- # appendSaveImg(6,getPlotImg())
- if(plotShow):
- plt.show()
- return thr1
-
-# from matplotlib.ticker import MaxNLocator
-# def plotArray(QVals, plotTitle, sort = False, plot=True ):
-# f, ax = plt.subplots()
-# if(sort):
-# QVals = sorted(QVals)
-# ax.bar(range(len(QVals)),QVals)
-# ax.set_title(plotTitle)
-# ax.set_ylabel("Values")
-# ax.set_xlabel("Position")
-# ax.xaxis.set_major_locator(MaxNLocator(integer=True))
-# if(plot):
-# plt.show()
-# # else: they will call this
-# # appendSaveImg(appendImgLvl,getPlotImg())
-
-
-def saveImg(path, final_marked):
- print('Saving Image to ' + path)
- cv2.imwrite(path, final_marked)
-
-
-def readResponse(template, image, name, savedir=None, autoAlign=False):
- global clahe
-
- try:
- img = image.copy()
- origDim = img.shape[:2]
- # print("noCropping dim", origDim)
- img = resize_util(img, template.dims[0], template.dims[1])
- # print("Resized dim", img.shape[:2])
-
- if(img.max() > img.min()):
- img = normalize_util(img)
- # Processing copies
- transp_layer = img.copy()
- final_marked = img.copy()
- # putLabel(final_marked,"Crop Size: " + str(origDim[0])+"x"+str(origDim[1]) + " "+name, size=1)
-
- morph = img.copy()
- appendSaveImg(3, morph)
-
- # TODO: evaluate if CLAHE is really req
- if(autoAlign):
- # Note: clahe is good for morphology, bad for thresholding
- morph = clahe.apply(morph)
- appendSaveImg(3, morph)
- # Remove shadows further, make columns/boxes darker (less gamma)
- morph = adjust_gamma(morph, GAMMA_LOW)
- ret, morph = cv2.threshold(morph, 220, 220, cv2.THRESH_TRUNC)
- morph = normalize_util(morph)
- appendSaveImg(3, morph)
- if(showimglvl >= 4):
- show("morph1", morph, 0, 1)
-
- # Overlay Transparencies
- alpha = 0.65
- alpha1 = 0.55
-
- boxW, boxH = template.bubbleDims
- lang = ['E', 'H']
- OMRresponse = {}
-
- multimarked, multiroll = 0, 0
-
- # TODO Make this part useful for visualizing status checks
- # blackVals=[0]
- # whiteVals=[255]
-
- if(showimglvl >= 5):
- # "QTYPE_ROLL":[]}#,"QTYPE_MED":[]}
- allCBoxvals = {"Int": [], "Mcq": []}
- # ,"QTYPE_ROLL":[]}#,"QTYPE_MED":[]}
- qNums = {"Int": [], "Mcq": []}
-
- # Find Shifts for the QBlocks --> Before calculating threshold!
- if(autoAlign):
- # print("Begin Alignment")
- # Open : erode then dilate
- # Vertical kernel
- v_kernel = cv2.getStructuringElement(cv2.MORPH_RECT, (2, 10))
- morph_v = cv2.morphologyEx(
- morph, cv2.MORPH_OPEN, v_kernel, iterations=3)
- ret, morph_v = cv2.threshold(morph_v, 200, 200, cv2.THRESH_TRUNC)
- morph_v = 255 - normalize_util(morph_v)
-
- if(showimglvl >= 3):
- show("morphed_vertical", morph_v, 0, 1)
-
- # show("morph1",morph,0,1)
- # show("morphed_vertical",morph_v,0,1)
-
- appendSaveImg(3, morph_v)
-
- morphTHR = 60 # for Mobile images
- # morphTHR = 40 # for scan Images
- # best tuned to 5x5 now
- _, morph_v = cv2.threshold(
- morph_v, morphTHR, 255, cv2.THRESH_BINARY)
- morph_v = cv2.erode(
- morph_v, np.ones(
- (5, 5), np.uint8), iterations=2)
-
- appendSaveImg(3, morph_v)
- # h_kernel = cv2.getStructuringElement(cv2.MORPH_RECT, (10, 2))
- # morph_h = cv2.morphologyEx(morph, cv2.MORPH_OPEN, h_kernel, iterations=3)
- # ret, morph_h = cv2.threshold(morph_h,200,200,cv2.THRESH_TRUNC)
- # morph_h = 255 - normalize_util(morph_h)
- # show("morph_h",morph_h,0,1)
- # _, morph_h = cv2.threshold(morph_h,morphTHR,255,cv2.THRESH_BINARY)
- # morph_h = cv2.erode(morph_h, np.ones((5,5),np.uint8), iterations = 2)
- if(showimglvl >= 3):
- show("morph_thr_eroded", morph_v, 0, 1)
-
- appendSaveImg(6, morph_v)
-
- # template alignment code
- # OUTPUT : each QBlock.shift is updated
- for QBlock in template.QBlocks:
- s, d = QBlock.orig, QBlock.dims
- # internal constants - wont need change much
- # TODO - ALIGN_STRIDE would depend on template's Dimensions
- ALIGN_STRIDE, MATCH_COL, ALIGN_STEPS = 1, 5, int(boxW * 2 / 3)
- shift, steps = 0, 0
- THK = 3
- while steps < ALIGN_STEPS:
- L = np.mean(morph_v[s[1]:s[1]+d[1],s[0]+shift-THK:-THK+s[0]+shift+MATCH_COL])
- R = np.mean(morph_v[s[1]:s[1]+d[1],s[0]+shift-MATCH_COL+d[0]+THK:THK+s[0]+shift+d[0]])
-
- # For demonstration purposes-
- if(QBlock.key == "Int1"):
- ret = morph_v.copy()
- cv2.rectangle(ret,(s[0]+shift-THK,s[1]),(s[0]+shift+THK+d[0],s[1]+d[1]),CLR_WHITE,3)
- appendSaveImg(6, ret)
- # print(shift, L, R)
- LW, RW = L > 100, R > 100
- if(LW):
- if(RW):
- break
- else:
- shift -= ALIGN_STRIDE
- else:
- if(RW):
- shift += ALIGN_STRIDE
- else:
- break
- steps += 1
-
- QBlock.shift = shift
- # print("Aligned QBlock: ",QBlock.key,"Corrected Shift:", QBlock.shift,", Dimensions:", QBlock.dims, "orig:", QBlock.orig,'\n')
- # print("End Alignment")
-
- final_align = None
- if(showimglvl >= 2):
- initial_align = drawTemplateLayout(img, template, shifted=False)
- final_align = drawTemplateLayout(
- img, template, shifted=True, draw_qvals=True)
- # appendSaveImg(4,mean_vals)
- appendSaveImg(2, initial_align)
- appendSaveImg(2, final_align)
- appendSaveImg(5, img)
- if(autoAlign):
- final_align = np.hstack((initial_align, final_align))
-
- # Get mean vals n other stats
- allQVals, allQStripArrs, allQStdVals = [], [], []
- totalQStripNo = 0
- for QBlock in template.QBlocks:
- QStdVals = []
- for qStrip, qBoxPts in QBlock.traverse_pts:
- QStripvals = []
- for pt in qBoxPts:
- # shifted
- x, y = (pt.x + QBlock.shift, pt.y)
- rect = [y, y + boxH, x, x + boxW]
- QStripvals.append(
- cv2.mean(img[rect[0]:rect[1], rect[2]:rect[3]])[0])
- QStdVals.append(round(np.std(QStripvals), 2))
- allQStripArrs.append(QStripvals)
- # _, _, _ = getGlobalThreshold(QStripvals, "QStrip Plot", plotShow=False, sortInPlot=True)
- # hist = getPlotImg()
- # show("QStrip "+qBoxPts[0].qNo, hist, 0, 1)
- allQVals.extend(QStripvals)
- # print(totalQStripNo, qBoxPts[0].qNo, QStdVals[len(QStdVals)-1])
- totalQStripNo += 1
- allQStdVals.extend(QStdVals)
- # print("Begin getGlobalThresholdStd")
- globalStdTHR, jstd_low, jstd_high = getGlobalThreshold(allQStdVals)# , "Q-wise Std-dev Plot", plotShow=True, sortInPlot=True)
- # print("End getGlobalThresholdStd")
- # print("Begin getGlobalThreshold")
- # plt.show()
- # hist = getPlotImg()
- # show("StdHist", hist, 0, 1)
-
- # Note: Plotting takes Significant times here --> Change Plotting args
- # to support showimglvl
- # , "Mean Intensity Histogram",plotShow=True, sortInPlot=True)
- globalTHR, j_low, j_high = getGlobalThreshold(allQVals, looseness=4)
-
- # TODO colorama
- print(
- "Thresholding:\t\t globalTHR: ", round(
- globalTHR, 2), "\tglobalStdTHR: ", round(
- globalStdTHR, 2), "\t(Looks like a Xeroxed OMR)" if(
- globalTHR == 255) else "")
- # plt.show()
- # hist = getPlotImg()
- # show("StdHist", hist, 0, 1)
-
- # print("End getGlobalThreshold")
-
- # if(showimglvl>=1):
- # hist = getPlotImg()
- # show("Hist", hist, 0, 1)
- # appendSaveImg(4,hist)
- # appendSaveImg(5,hist)
- # appendSaveImg(2,hist)
-
- perOMRThresholdAvg, totalQStripNo, totalQBoxNo = 0, 0, 0
- for QBlock in template.QBlocks:
- blockQStripNo = 1 # start from 1 is fine here
- shift = QBlock.shift
- s, d = QBlock.orig, QBlock.dims
- key = QBlock.key[:3]
- # cv2.rectangle(final_marked,(s[0]+shift,s[1]),(s[0]+shift+d[0],s[1]+d[1]),CLR_BLACK,3)
- for qStrip, qBoxPts in QBlock.traverse_pts:
- # All Black or All White case
- noOutliers = allQStdVals[totalQStripNo] < globalStdTHR
- # print(totalQStripNo, qBoxPts[0].qNo, allQStdVals[totalQStripNo], "noOutliers:", noOutliers)
- perQStripThreshold = getLocalThreshold(qBoxPts[0].qNo, allQStripArrs[totalQStripNo],
- globalTHR, noOutliers,
- "Mean Intensity Histogram for " + key + "." +
- qBoxPts[0].qNo + '.' +
- str(blockQStripNo),
- showimglvl >= 6)
- # print(qBoxPts[0].qNo,key,blockQStripNo, "THR: ",round(perQStripThreshold,2))
- perOMRThresholdAvg += perQStripThreshold
-
- # Note: Little debugging visualization - view the particular Qstrip
- # if(
- # 0
- # # or "q17" in (qBoxPts[0].qNo)
- # # or (qBoxPts[0].qNo+str(blockQStripNo))=="q15"
- # ):
- # st, end = qStrip
- # show("QStrip: "+key+"-"+str(blockQStripNo), img[st[1] : end[1], st[0]+shift : end[0]+shift],0)
-
- for pt in qBoxPts:
- # shifted
- x, y = (pt.x + QBlock.shift, pt.y)
- boxval0 = allQVals[totalQBoxNo]
- detected = perQStripThreshold > boxval0
-
- # TODO: add an option to select PLUS SIGN RESPONSE READING
- # extra_check_rects = []
- # # [y,y+boxH,x,x+boxW]
- # for rect in extra_check_rects:
- # # Note: This is NOT pixel-based thresholding, It is boxed mean-thresholding
- # boxval = cv2.mean(img[ rect[0]:rect[1] , rect[2]:rect[3] ])[0]
- # if(perQStripThreshold > boxval):
- # # for critical analysis
- # boxval0 = max(boxval,boxval0)
- # detected=True
- # break
-
- if (detected):
- cv2.rectangle(final_marked,(int(x+boxW/12),int(y+boxH/12)),(int(x+boxW-boxW/12),int(y+boxH-boxH/12)), CLR_DARK_GRAY, 3)
- else:
- cv2.rectangle(final_marked,(int(x+boxW/10),int(y+boxH/10)),(int(x+boxW-boxW/10),int(y+boxH-boxH/10)), CLR_GRAY,-1)
-
- # TODO Make this part useful! (Abstract visualizer to check
- # status)
- if (detected):
- q, val = pt.qNo, str(pt.val)
- cv2.putText(
- final_marked, val, (x, y), cv2.FONT_HERSHEY_SIMPLEX, TEXT_SIZE, (20, 20, 10), int(
- 1 + 3.5 * TEXT_SIZE))
- # Only send rolls multi-marked in the directory
- multimarkedL = q in OMRresponse
- multimarked = multimarkedL or multimarked
- OMRresponse[q] = (
- OMRresponse[q] + val) if multimarkedL else val
- multiroll = multimarkedL and 'roll' in str(q)
- # blackVals.append(boxval0)
- # else:
- # whiteVals.append(boxval0)
-
- totalQBoxNo += 1
- # /for qBoxPts
- # /for qStrip
-
- if(showimglvl >= 5):
- if(key in allCBoxvals):
- qNums[key].append(key[:2] + '_c' + str(blockQStripNo))
- allCBoxvals[key].append(allQStripArrs[totalQStripNo])
-
- blockQStripNo += 1
- totalQStripNo += 1
- # /for QBlock
-
- # TODO: move this validation into template.py -
- if(totalQStripNo == 0):
- print(
- "\n\t UNEXPECTED Template Incorrect Error: totalQStripNo is zero! QBlocks: ",
- template.QBlocks)
- exit(21)
-
- perOMRThresholdAvg /= totalQStripNo
- perOMRThresholdAvg = round(perOMRThresholdAvg, 2)
- # Translucent
- cv2.addWeighted(
- final_marked,
- alpha,
- transp_layer,
- 1 - alpha,
- 0,
- final_marked)
- # Box types
- if(showimglvl >= 5):
- # plt.draw()
- f, axes = plt.subplots(len(allCBoxvals), sharey=True)
- f.canvas.set_window_title(name)
- ctr = 0
- typeName = {
- "Int": "Integer",
- "Mcq": "MCQ",
- "Med": "MED",
- "Rol": "Roll"}
- for k, boxvals in allCBoxvals.items():
- axes[ctr].title.set_text(typeName[k] + " Type")
- axes[ctr].boxplot(boxvals)
- # thrline=axes[ctr].axhline(perOMRThresholdAvg,color='red',ls='--')
- # thrline.set_label("Average THR")
- axes[ctr].set_ylabel("Intensity")
- axes[ctr].set_xticklabels(qNums[k])
- # axes[ctr].legend()
- ctr += 1
- # imshow will do the waiting
- plt.tight_layout(pad=0.5)
- plt.show()
-
- if(showimglvl >= 3 and final_align is not None):
- final_align = resize_util_h(final_align, int(display_height))
- # [final_align.shape[1],0])
- show("Template Alignment Adjustment", final_align, 0, 0)
-
- # TODO: refactor "type(savedir) != type(None) "
- if (saveMarked and type(savedir) != type(None)):
- if(multiroll):
- savedir = savedir + '_MULTI_/'
- saveImg(savedir + name, final_marked)
-
- if(showimglvl >= 1):
- show("Final Marked Bubbles : " + name,
- resize_util_h(final_marked, int(display_height * 1.3)), 1, 1)
-
- appendSaveImg(2, final_marked)
-
- for i in range(saveimglvl):
- saveOrShowStacks(i + 1, name, savedir)
-
- return OMRresponse, final_marked, multimarked, multiroll
-
- except Exception as e:
- exc_type, exc_obj, exc_tb = sys.exc_info()
- fname = os.path.split(exc_tb.tb_frame.f_code.co_filename)[1]
- print("Error from readResponse: ", e)
- print(exc_type, fname, exc_tb.tb_lineno)
-
-
-def saveOrShowStacks(key, name, savedir=None, pause=1):
- global saveImgList
- if(saveimglvl >= int(key) and saveImgList[key] != []):
- result = np.hstack(
- tuple([resize_util_h(img, uniform_height) for img in saveImgList[key]]))
- result = resize_util(result,
- min(len(saveImgList[key]) * uniform_width // 3,
- int(uniform_width * 2.5)))
- if (type(savedir) != type(None)):
- saveImg(savedir+'stack/'+name+'_'+str(key)+'_stack.jpg', result)
- else:
- show(name + '_' + str(key), result, pause, 0)




 +
+
+
+ [](https://www.paypal.me/Udayraj123/500)
+
+_Find OMRChecker on_ [**_Product Hunt_**](https://www.producthunt.com/posts/omr-checker/) **|** [**_Reddit_**](https://www.reddit.com/r/computervision/comments/ccbj6f/omrchecker_grade_exams_using_python_and_opencv/) **|** [**Discord**](https://discord.gg/qFv2Vqf) **|** [**Linkedin**](https://www.linkedin.com/pulse/open-source-talks-udayraj-udayraj-deshmukh/) **|** [**goodfirstissue.dev**](https://goodfirstissue.dev/language/python) **|** [**codepeak.tech**](https://www.codepeak.tech/) **|** [**fossoverflow.dev**](https://fossoverflow.dev/projects) **|** [**Interview on Console by CodeSee**](https://console.substack.com/p/console-140) **|** [**Open Source Hub**](https://opensourcehub.io/udayraj123/omrchecker)
-
-*Find OMRChecker on* [***Product Hunt***](https://www.producthunt.com/posts/omr-checker/) **|** [***Hacker News***](https://news.ycombinator.com/item?id=20420602) **|** [***Reddit***](https://www.reddit.com/r/computervision/comments/ccbj6f/omrchecker_grade_exams_using_python_and_opencv/) **|** [***Swyya***](https://www.swyya.com/projects/omrchecker) **|** [](https://discord.gg/qFv2Vqf)
+
+
diff --git a/docs/assets/colored_output.jpg b/docs/assets/colored_output.jpg
new file mode 100644
index 00000000..3cafa473
Binary files /dev/null and b/docs/assets/colored_output.jpg differ
diff --git a/globals.py b/globals.py
deleted file mode 100644
index 5f9cd929..00000000
--- a/globals.py
+++ /dev/null
@@ -1,140 +0,0 @@
-"""
-
-Designed and Developed by-
-Udayraj Deshmukh
-https://github.com/Udayraj123
-
-"""
-
-"""
-Constants
-"""
-display_height = int(480)
-display_width = int(640)
-windowWidth = 1280
-windowHeight = 720
-
-saveMarked = 1
-saveCropped = 1
-showimglvl = 4
-saveimglvl = 0
-PRELIM_CHECKS = 0
-saveImgList = {}
-resetpos = [0, 0]
-explain = 0
-# autorotate=1
-
-BATCH_NO = 1000
-NO_MARKER_ERR = 12
-MULTI_BUBBLE_WARN = 15
-
-# name of template file
-TEMPLATE_FILE = 'template.json'
-MARKER_FILE = "omr_marker.jpg"
-
-# For preProcessing
-GAMMA_LOW = 0.7
-GAMMA_HIGH = 1.25
-
-ERODE_SUB_OFF = 1
-
-# For new ways of determining threshold
-MIN_GAP, MIN_STD = 30, 25
-MIN_JUMP = 25
-# If only not confident, take help of globalTHR
-CONFIDENT_JUMP = MIN_JUMP + 15
-JUMP_DELTA = 30
-# MIN_GAP : worst case gap of black and gray
-
-# Templ alignment parameters
-ALIGN_RANGE = range(-5, 6, 1)
-# TODO ^THIS SHOULD BE IN LAYOUT FILE AS ITS RELATED TO DIMENSIONS
-# ALIGN_RANGE = [-6,-4,-2,-1,0,1,2,4,6]
-
-# max threshold difference for template matching
-thresholdVar = 0.41
-
-# TODO: remove unnec variables here-
-thresholdCircle = 0.3
-marker_rescale_range = (35, 100)
-marker_rescale_steps = 10
-
-# Presentation variables
-uniform_height = int(1231 / 1.5)
-uniform_width = int(1000 / 1.5)
-# Original dims are about (3527, 2494)
-
-# Any input images should be resized to this--
-uniform_width_hd = int(uniform_width * 1.5)
-uniform_height_hd = int(uniform_height * 1.5)
-
-TEXT_SIZE = 0.95
-CLR_BLACK = (50, 150, 150)
-CLR_WHITE = (250, 250, 250)
-CLR_GRAY = (130, 130, 130)
-# CLR_DARK_GRAY = (190,190,190)
-CLR_DARK_GRAY = (100, 100, 100)
-
-MIN_PAGE_AREA = 80000
-
-# Filepaths
-
-
-class Paths:
- def __init__(self, output):
- self.output = output
- self.saveMarkedDir = f'{output}/CheckedOMRs/'
- self.resultDir = f'{output}/Results/'
- self.manualDir = f'{output}/Manual/'
- self.errorsDir = f'{self.manualDir}ErrorFiles/'
- self.badRollsDir = f'{self.manualDir}BadRollNosFiles/'
- self.multiMarkedDir = f'{self.manualDir}MultiMarkedFiles/'
-
-
-"""
-Variables
-"""
-filesMoved = 0
-filesNotMoved = 0
-
-# for positioning image windows
-windowX, windowY = 0, 0
-
-
-# TODO: move to template or similar json
-Answers = {
- 'J': {
- 'q1': ['B'], 'q2': ['B'], 'q3': ['B'], 'q4': ['C'], 'q5': ['0', '00'], 'q6': ['0', '00'], 'q7': ['4', '04'],
- 'q8': ['9', '09'], 'q9': ['11', '11'], 'q10': ['C'], 'q11': ['C'], 'q12': ['B'], 'q13': ['C'],
- 'q14': ['C'], 'q15': ['B'], 'q16': ['C'], 'q17': ['BONUS'], 'q18': ['A'], 'q19': ['C'], 'q20': ['B']},
- 'H': {
- 'q1': ['B'], 'q2': ['BONUS'], 'q3': ['A'], 'q4': ['B'], 'q5': ['A'], 'q6': ['B'], 'q7': ['B'],
- 'q8': ['C'], 'q9': ['4', '04'], 'q10': ['4', '04'], 'q11': ['5', '05'], 'q12': ['1', '01'], 'q13': ['28'],
- 'q14': ['C'], 'q15': ['B'], 'q16': ['C'], 'q17': ['C'], 'q18': ['C'], 'q19': ['B'], 'q20': ['C']},
- 'JK': {
- 'q1': ['B'], 'q2': ['B'], 'q3': ['B'], 'q4': ['C'], 'q5': ['0', '00'], 'q6': ['0', '00'], 'q7': ['4', '04'],
- 'q8': ['9', '09'], 'q9': ['11', '11'], 'q10': ['C'], 'q11': ['C'], 'q12': ['B'], 'q13': ['C'],
- 'q14': ['C'], 'q15': ['B'], 'q16': ['C'], 'q17': ['BONUS'], 'q18': ['A'], 'q19': ['C'], 'q20': ['B']},
- 'HK': {
- 'q1': ['B'], 'q2': ['BONUS'], 'q3': ['A'], 'q4': ['B'], 'q5': ['B'], 'q6': ['B'], 'q7': ['B'],
- 'q8': ['C'], 'q9': ['4', '04'], 'q10': ['4', '04'], 'q11': ['5', '05'], 'q12': ['1', '01'], 'q13': ['28'],
- 'q14': ['C'], 'q15': ['B'], 'q16': ['C'], 'q17': ['C'], 'q18': ['C'], 'q19': ['B'], 'q20': ['C']},
-}
-
-# TODO: Make this generalized and move it to samples
-Sections = {
- 'J': {
- 'Fibo1': {'ques': [1, 2, 3, 4], '+seq': [2, 3, 5, 8], '-seq': [0, 1, 1, 2]},
- 'Power1': {'ques': [5, 6, 7, 8, 9], '+seq': [1, 2, 4, 8, 16], '-seq': [0, 0, 0, 0, 0]},
- 'Fibo2': {'ques': [10, 11, 12, 13], '+seq': [2, 3, 5, 8], '-seq': [0, 1, 1, 2]},
- 'allNone1': {'ques': [14, 15, 16], 'marks': 12},
- 'Boom1': {'ques': [17, 18, 19, 20], '+seq': [3, 3, 3, 3], '-seq': [1, 1, 1, 1]},
- },
- 'H': {
- 'Boom1': {'ques': [1, 2, 3, 4], '+seq': [3, 3, 3, 3], '-seq': [1, 1, 1, 1]},
- 'Fibo1': {'ques': [5, 6, 7, 8], '+seq': [2, 3, 5, 8], '-seq': [0, 1, 1, 2]},
- 'Power1': {'ques': [9, 10, 11, 12, 13], '+seq': [1, 2, 4, 8, 16], '-seq': [0, 0, 0, 0, 0]},
- 'allNone1': {'ques': [14, 15, 16], 'marks': 12},
- 'Boom2': {'ques': [17, 18, 19, 20], '+seq': [3, 3, 3, 3], '-seq': [1, 1, 1, 1]},
- },
-}
diff --git a/main.py b/main.py
index eac70a50..dfecbca3 100644
--- a/main.py
+++ b/main.py
@@ -1,546 +1,99 @@
"""
-Designed and Developed by-
-Udayraj Deshmukh
-https://github.com/Udayraj123
+ OMRChecker
+
+ Author: Udayraj Deshmukh
+ Github: https://github.com/Udayraj123
"""
-import re
-import os
-import cv2
import argparse
-import numpy as np
-import pandas as pd
-import matplotlib.pyplot as plt
-
-# from utils import * #Now imported via template
-from globals import *
-from template import *
-from glob import glob
-from csv import QUOTE_NONNUMERIC
-from time import localtime, strftime, time
-
-
-# TODO(beginner task) :-
-# from colorama import init
-# init()
-# from colorama import Fore, Back, Style
-
-def process_dir(root_dir, subdir, template):
- curr_dir = os.path.join(root_dir, subdir)
-
- # Look for template in current dir
- template_file = os.path.join(curr_dir, TEMPLATE_FILE)
- if os.path.exists(template_file):
- template = Template(template_file)
-
- # look for images in current dir to process
- paths = Paths(os.path.join(args['output_dir'], subdir))
- exts = ('*.png', '*.jpg')
- omr_files = sorted(
- [f for ext in exts for f in glob(os.path.join(curr_dir, ext))])
-
- # Exclude marker image if exists
- if(template and template.marker_path):
- omr_files = [f for f in omr_files if f != template.marker_path]
-
- subfolders = sorted([file for file in os.listdir(
- curr_dir) if os.path.isdir(os.path.join(curr_dir, file))])
- if omr_files:
- args_local = args.copy()
- if("OverrideFlags" in template.options):
- args_local.update(template.options["OverrideFlags"])
- print('\n------------------------------------------------------------------')
- print(f'Processing directory "{curr_dir}" with settings- ')
- print("\tTotal images : %d" % (len(omr_files)))
- print("\tCropping Enabled : " + str(not args_local["noCropping"]))
- print("\tAuto Alignment : " + str(args_local["autoAlign"]))
- print("\tUsing Template : " + str(template.path) if(template) else "N/A")
- print("\tUsing Marker : " + str(template.marker_path)
- if(template.marker is not None) else "N/A")
- print('')
-
- if not template:
- print(f'Error: No template file when processing {curr_dir}.')
- print(
- f' Place {TEMPLATE_FILE} in the directory or specify a template using -t.')
- return
-
- setup_dirs(paths)
- output_set = setup_output(paths, template)
- process_files(omr_files, template, args_local, output_set)
- elif(len(subfolders) == 0):
- # the directory should have images or be non-leaf
- print(f'Note: No valid images or subfolders found in {curr_dir}')
-
- # recursively process subfolders
- for folder in subfolders:
- process_dir(root_dir, os.path.join(subdir, folder), template)
-
-
-def checkAndMove(error_code, filepath, filepath2):
- # print("Dummy Move: "+filepath, " --> ",filepath2)
- global filesNotMoved
- filesNotMoved += 1
- return True
-
- global filesMoved
- if(not os.path.exists(filepath)):
- print('File already moved')
- return False
- if(os.path.exists(filepath2)):
- print('ERROR : Duplicate file at ' + filepath2)
- return False
-
- print("Moved: " + filepath, " --> ", filepath2)
- os.rename(filepath, filepath2)
- filesMoved += 1
- return True
-
-
-def processOMR(template, omrResp):
- # Note: This is a reference function. It is not part of the OMR checker
- # So its implementation is completely subjective to user's requirements.
- csvResp = {}
-
- # symbol for absent response
- UNMARKED_SYMBOL = ''
-
- # print("omrResp",omrResp)
-
- # Multi-column/multi-row questions which need to be concatenated
- for qNo, respKeys in template.concats.items():
- csvResp[qNo] = ''.join([omrResp.get(k, UNMARKED_SYMBOL)
- for k in respKeys])
-
- # Single-column/single-row questions
- for qNo in template.singles:
- csvResp[qNo] = omrResp.get(qNo, UNMARKED_SYMBOL)
-
- # Note: Concatenations and Singles together should be mutually exclusive
- # and should cover all questions in the template(exhaustive)
- # TODO: ^add a warning if omrResp has unused keys remaining
- return csvResp
-
-
-def report(
- Status,
- streak,
- scheme,
- qNo,
- marked,
- ans,
- prevmarks,
- currmarks,
- marks):
- print(
- '%s \t %s \t\t %s \t %s \t %s \t %s \t %s ' % (qNo,
- Status,
- str(streak),
- '[' + scheme + '] ',
- (str(prevmarks) + ' + ' + str(currmarks) + ' =' + str(marks)),
- str(marked),
- str(ans)))
-
-# check sectionwise only.
-
-
-def evaluate(resp, squad="H", explain=False):
- # TODO: @contributors - Need help generalizing this function
- global Answers, Sections
- marks = 0
- answers = Answers[squad]
- if(explain):
- print('Question\tStatus \t Streak\tSection \tMarks_Update\tMarked:\tAnswer:')
- for scheme, section in Sections[squad].items():
- sectionques = section['ques']
- prevcorrect = None
- allflag = 1
- streak = 0
- for q in sectionques:
- qNo = 'q' + str(q)
- ans = answers[qNo]
- marked = resp.get(qNo, 'X')
- firstQ = sectionques[0]
- lastQ = sectionques[len(sectionques) - 1]
- unmarked = marked == 'X' or marked == ''
- bonus = 'BONUS' in ans
- correct = bonus or (marked in ans)
- inrange = 0
-
- if(unmarked or int(q) == firstQ):
- streak = 0
- elif(prevcorrect == correct):
- streak += 1
- else:
- streak = 0
-
- if('allNone' in scheme):
- # loop on all sectionques
- allflag = allflag and correct
- if(q == lastQ):
- # at the end check allflag
- prevcorrect = correct
- currmarks = section['marks'] if allflag else 0
- else:
- currmarks = 0
-
- elif('Proxy' in scheme):
- a = int(ans[0])
- # proximity check
- inrange = 1 if unmarked else (
- float(abs(int(marked) - a)) / float(a) <= 0.25)
- currmarks = section['+marks'] if correct else (
- 0 if inrange else -section['-marks'])
-
- elif('Fibo' in scheme or 'Power' in scheme or 'Boom' in scheme):
- currmarks = section['+seq'][streak] if correct else (
- 0 if unmarked else -section['-seq'][streak])
- elif('TechnoFin' in scheme):
- currmarks = 0
- else:
- print('Invalid Sections')
- prevmarks = marks
- marks += currmarks
-
- if(explain):
- if bonus:
- report('BonusQ', streak, scheme, qNo, marked,
- ans, prevmarks, currmarks, marks)
- elif correct:
- report('Correct', streak, scheme, qNo, marked,
- ans, prevmarks, currmarks, marks)
- elif unmarked:
- report('Unmarked', streak, scheme, qNo, marked,
- ans, prevmarks, currmarks, marks)
- elif inrange:
- report('InProximity', streak, scheme, qNo,
- marked, ans, prevmarks, currmarks, marks)
- else:
- report('Incorrect', streak, scheme, qNo,
- marked, ans, prevmarks, currmarks, marks)
-
- prevcorrect = correct
-
- return marks
-
-
-def setup_output(paths, template):
- ns = argparse.Namespace()
- print("\nChecking Files...")
-
- # Include current output paths
- ns.paths = paths
-
- # custom sort: To use integer order in question names instead of
- # alphabetical - avoids q1, q10, q2 and orders them q1, q2, ..., q10
- ns.respCols = sorted(list(template.concats.keys()) + template.singles,
- key=lambda x: int(x[1:]) if ord(x[1]) in range(48, 58) else 0)
- ns.emptyResp = [''] * len(ns.respCols)
- ns.sheetCols = ['file_id', 'input_path',
- 'output_path', 'score'] + ns.respCols
- ns.OUTPUT_SET = []
- ns.filesObj = {}
- ns.filesMap = {
- "Results": paths.resultDir + 'Results_' + timeNowHrs + '.csv',
- "MultiMarked": paths.manualDir + 'MultiMarkedFiles_.csv',
- "Errors": paths.manualDir + 'ErrorFiles_.csv',
- "BadRollNos": paths.manualDir + 'BadRollNoFiles_.csv'
- }
-
- for fileKey, fileName in ns.filesMap.items():
- if(not os.path.exists(fileName)):
- print("Note: Created new file: %s" % (fileName))
- # still append mode req [THINK!]
- ns.filesObj[fileKey] = open(fileName, 'a')
- # Create Header Columns
- pd.DataFrame([ns.sheetCols], dtype=str).to_csv(
- ns.filesObj[fileKey], quoting=QUOTE_NONNUMERIC, header=False, index=False)
- else:
- print('Present : appending to %s' % (fileName))
- ns.filesObj[fileKey] = open(fileName, 'a')
-
- return ns
-
-
-''' TODO: Refactor into new process flow.
- Currently I have no idea what this does so I left it out'''
-
-
-def preliminary_check():
- filesCounter = 0
- # PRELIM_CHECKS for thresholding
- if(PRELIM_CHECKS):
- # TODO: add more using unit testing
- ALL_WHITE = 255 * \
- np.ones((TEMPLATE.dims[1], TEMPLATE.dims[0]), dtype='uint8')
- OMRresponseDict, final_marked, MultiMarked, multiroll = readResponse(
- ALL_WHITE, name="ALL_WHITE", savedir=None, autoAlign=True)
- print("ALL_WHITE", OMRresponseDict)
- if(OMRresponseDict != {}):
- print("Preliminary Checks Failed.")
- exit(12)
- ALL_BLACK = np.zeros(
- (TEMPLATE.dims[1], TEMPLATE.dims[0]), dtype='uint8')
- OMRresponseDict, final_marked, MultiMarked, multiroll = readResponse(
- ALL_BLACK, name="ALL_BLACK", savedir=None, autoAlign=True)
- print("ALL_BLACK", OMRresponseDict)
- show("Confirm : All bubbles are black", final_marked, 1, 1)
-
-
-def process_files(omr_files, template, args, out):
- start_time = int(time())
- filesCounter = 0
- filesNotMoved = 0
-
- for filepath in omr_files:
- filesCounter += 1
- # For windows filesystem support: all '\' will be replaced by '/'
- filepath = filepath.replace(os.sep, '/')
-
- # Prefixing a 'r' to use raw string (escape character '\' is taken
- # literally)
- finder = re.search(r'.*/(.*)/(.*)', filepath, re.IGNORECASE)
- if(finder):
- inputFolderName, filename = finder.groups()
- else:
- print("Error: Filepath not matching to Regex: " + filepath)
- continue
- # set global var for reading
-
- inOMR = cv2.imread(filepath, cv2.IMREAD_GRAYSCALE)
- print(
- '\n[%d] Processing image: \t' %
- (filesCounter),
- filepath,
- "\tResolution: ",
- inOMR.shape)
-
- OMRCrop = getROI(inOMR, filename, noCropping=args["noCropping"])
-
- if(OMRCrop is None):
- # Error OMR - could not crop
- newfilepath = out.paths.errorsDir + filename
- out.OUTPUT_SET.append([filename] + out.emptyResp)
- if(checkAndMove(NO_MARKER_ERR, filepath, newfilepath)):
- err_line = [filename, filepath,
- newfilepath, "NA"] + out.emptyResp
- pd.DataFrame(
- err_line,
- dtype=str).T.to_csv(
- out.filesObj["Errors"],
- quoting=QUOTE_NONNUMERIC,
- header=False,
- index=False)
- continue
-
- if template.marker is not None:
- OMRCrop = handle_markers(OMRCrop, template.marker, filename)
-
- if(args["setLayout"]):
- templateLayout = drawTemplateLayout(
- OMRCrop, template, shifted=False, border=2)
- show("Template Layout", templateLayout, 1, 1)
- continue
-
- # uniquify
- file_id = inputFolderName + '_' + filename
- savedir = out.paths.saveMarkedDir
- OMRresponseDict, final_marked, MultiMarked, multiroll = \
- readResponse(template, OMRCrop, name=file_id,
- savedir=savedir, autoAlign=args["autoAlign"])
-
- # concatenate roll nos, set unmarked responses, etc
- resp = processOMR(template, OMRresponseDict)
- print("\nRead Response: \t", resp)
-
- # This evaluates and returns the score attribute
- score = evaluate(resp, explain=explain)
- respArray = []
- for k in out.respCols:
- respArray.append(resp[k])
-
- out.OUTPUT_SET.append([filename] + respArray)
-
- # TODO: Add roll number validation here
- if(MultiMarked == 0):
- filesNotMoved += 1
- newfilepath = savedir + file_id
- # Enter into Results sheet-
- results_line = [filename, filepath, newfilepath, score] + respArray
- # Write/Append to results_line file(opened in append mode)
- pd.DataFrame(
- results_line,
- dtype=str).T.to_csv(
- out.filesObj["Results"],
- quoting=QUOTE_NONNUMERIC,
- header=False,
- index=False)
- print("[%d] Graded with score: %.2f" %
- (filesCounter, score), '\t file_id: ', file_id)
- # print(filesCounter,file_id,resp['Roll'],'score : ',score)
- else:
- # MultiMarked file
- print('[%d] MultiMarked, moving File: %s' %
- (filesCounter, file_id))
- newfilepath = out.paths.multiMarkedDir + filename
- if(checkAndMove(MULTI_BUBBLE_WARN, filepath, newfilepath)):
- mm_line = [filename, filepath, newfilepath, "NA"] + respArray
- pd.DataFrame(
- mm_line,
- dtype=str).T.to_csv(
- out.filesObj["MultiMarked"],
- quoting=QUOTE_NONNUMERIC,
- header=False,
- index=False)
- # else:
- # TODO: Add appropriate record handling here
- # pass
-
- # flush after every 20 files for a live view
- if(filesCounter % 20 == 0 or filesCounter == len(omr_files)):
- for fileKey in out.filesMap.keys():
- out.filesObj[fileKey].flush()
-
- timeChecking = round(time() - start_time, 2) if filesCounter else 1
- print('')
- print('Total files moved : %d ' % (filesMoved))
- print('Total files not moved : %d ' % (filesNotMoved))
- print('------------------------------')
- print(
- 'Total files processed : %d (%s)' %
- (filesCounter,
- 'Sum Tallied!' if filesCounter == (
- filesMoved +
- filesNotMoved) else 'Not Tallying!'))
-
- if(showimglvl <= 0):
- print(
- '\nFinished Checking %d files in %.1f seconds i.e. ~%.1f minutes.' %
- (filesCounter, timeChecking, timeChecking / 60))
- print('OMR Processing Rate :\t ~ %.2f seconds/OMR' %
- (timeChecking / filesCounter))
- print('OMR Processing Speed :\t ~ %.2f OMRs/minute' %
- ((filesCounter * 60) / timeChecking))
- else:
- print("\nTotal script time :", timeChecking, "seconds")
-
- if(showimglvl <= 1):
- # TODO: colorama this
- print(
- "\nTip: To see some awesome visuals, open globals.py and increase 'showimglvl'")
-
- evaluate_correctness(template, out)
-
- # Use this data to train as +ve feedback
- if(showimglvl >= 0 and filesCounter > 10):
- # TODO: Find good parameters to plot and depict image set quality
- for x in [thresholdCircles]: # ,badThresholds,veryBadPoints, , mbs]:
- if(x != []):
- x = pd.DataFrame(x)
- print(x.describe())
- plt.plot(range(len(x)), x)
- plt.title("Mystery Plot")
- plt.show()
- else:
- print(x)
-
-
-# Evaluate accuracy based on OMRDataset file generated through moderation
-# portal on the same set of images
-def evaluate_correctness(template, out):
- # TODO: TEST_FILE WOULD BE RELATIVE TO INPUT SUBDIRECTORY NOW-
- TEST_FILE = 'inputs/OMRDataset.csv'
- if(os.path.exists(TEST_FILE)):
- print("\nStarting evaluation for: " + TEST_FILE)
-
- TEST_COLS = ['file_id'] + out.respCols
- y_df = pd.read_csv(
- TEST_FILE, dtype=str)[TEST_COLS].replace(
- np.nan, '', regex=True).set_index('file_id')
-
- if(np.any(y_df.index.duplicated)):
- y_df_filtered = y_df.loc[~y_df.index.duplicated(keep='first')]
- print(
- "WARNING: Found duplicate File-ids in file %s. Removed %d rows from testing data. Rows remaining: %d" %
- (TEST_FILE, y_df.shape[0] - y_df_filtered.shape[0], y_df_filtered.shape[0]))
- y_df = y_df_filtered
-
- x_df = pd.DataFrame(
- out.OUTPUT_SET,
- dtype=str,
- columns=TEST_COLS).set_index('file_id')
- # print("x_df",x_df.head())
- # print("\ny_df",y_df.head())
- intersection = y_df.index.intersection(x_df.index)
-
- # Checking if the merge is okay
- if(intersection.size == x_df.index.size):
- y_df = y_df.loc[intersection]
- x_df['TestResult'] = (x_df == y_df).all(axis=1).astype(int)
- print(x_df.head())
- print("\n\t Accuracy on the %s Dataset: %.6f" %
- (TEST_FILE, (x_df['TestResult'].sum() / x_df.shape[0])))
- else:
- print(
- "\nERROR: Insufficient Testing Data: Have you appended MultiMarked data yet?")
- print("Missing File-ids: ",
- list(x_df.index.difference(intersection)))
-
-
-timeNowHrs = strftime("%I%p", localtime())
-
-# construct the argument parse and parse the arguments
-argparser = argparse.ArgumentParser()
-# https://docs.python.org/3/howto/argparse.html
-# store_true: if the option is specified, assign the value True to
-# args.verbose. Not specifying it implies False.
-argparser.add_argument(
- "-c",
- "--noCropping",
- required=False,
- dest='noCropping',
- action='store_true',
- help="Disables page contour detection - used when page boundary is not visible e.g. document scanner.")
-argparser.add_argument(
- "-a",
- "--autoAlign",
- required=False,
- dest='autoAlign',
- action='store_true',
- help="(experimental) Enables automatic template alignment - use if the scans show slight misalignments.")
-argparser.add_argument(
- "-l",
- "--setLayout",
- required=False,
- dest='setLayout',
- action='store_true',
- help="Set up OMR template layout - modify your json file and run again until the template is set.")
-argparser.add_argument("-i", "--inputDir", required=False, action='append',
- dest='input_dir', help="Specify an input directory.")
-argparser.add_argument("-o", "--outputDir", default='outputs', required=False,
- dest='output_dir', help="Specify an output directory.")
-argparser.add_argument(
- "-t",
- "--template",
- required=False,
- dest='template',
- help="Specify a default template if no template file in input directories.")
-
-
-args, unknown = argparser.parse_known_args()
-args = vars(args)
-if(len(unknown) > 0):
- print("\nError: Unknown arguments:", unknown)
- argparser.print_help()
- exit(11)
-
-if args['template']:
- args['template'] = Template(args['template'])
-
-if args['input_dir'] is None:
- args['input_dir'] = ['inputs']
-
-for root in args['input_dir']:
- process_dir(root, '', args['template'])
+import sys
+from pathlib import Path
+
+from src.entry import entry_point
+from src.logger import logger
+
+
+def parse_args():
+ # construct the argument parse and parse the arguments
+ argparser = argparse.ArgumentParser()
+
+ argparser.add_argument(
+ "-i",
+ "--inputDir",
+ default=["inputs"],
+ # https://docs.python.org/3/library/argparse.html#nargs
+ nargs="*",
+ required=False,
+ type=str,
+ dest="input_paths",
+ help="Specify an input directory.",
+ )
+
+ argparser.add_argument(
+ "-d",
+ "--debug",
+ required=False,
+ dest="debug",
+ action="store_false",
+ help="Enables debugging mode for showing detailed errors",
+ )
+
+ argparser.add_argument(
+ "-o",
+ "--outputDir",
+ default="outputs",
+ required=False,
+ dest="output_dir",
+ help="Specify an output directory.",
+ )
+
+ argparser.add_argument(
+ "-a",
+ "--autoAlign",
+ required=False,
+ dest="autoAlign",
+ action="store_true",
+ help="(experimental) Enables automatic template alignment - \
+ use if the scans show slight misalignments.",
+ )
+
+ argparser.add_argument(
+ "-l",
+ "--setLayout",
+ required=False,
+ dest="setLayout",
+ action="store_true",
+ help="Set up OMR template layout - modify your json file and \
+ run again until the template is set.",
+ )
+
+ (
+ args,
+ unknown,
+ ) = argparser.parse_known_args()
+
+ args = vars(args)
+
+ if len(unknown) > 0:
+ logger.warning(f"\nError: Unknown arguments: {unknown}", unknown)
+ argparser.print_help()
+ exit(11)
+ return args
+
+
+def entry_point_for_args(args):
+ if args["debug"] is True:
+ # Disable tracebacks
+ sys.tracebacklimit = 0
+ for root in args["input_paths"]:
+ entry_point(
+ Path(root),
+ args,
+ )
+
+
+if __name__ == "__main__":
+ args = parse_args()
+ entry_point_for_args(args)
diff --git a/pyproject.toml b/pyproject.toml
new file mode 100644
index 00000000..060309be
--- /dev/null
+++ b/pyproject.toml
@@ -0,0 +1,18 @@
+[tool.black]
+exclude = '''
+(
+ /(
+ \.eggs # exclude a few common directories in the
+ | \.git # root of the project
+ | \.venv
+ | _build
+ | build
+ | dist
+ )/
+ | foo.py # also separately exclude a file named foo.py in
+ # the root of the project
+)
+'''
+include = '\.pyi?$'
+line-length = 88
+target-version = ['py37']
diff --git a/pytest.ini b/pytest.ini
new file mode 100644
index 00000000..84008a21
--- /dev/null
+++ b/pytest.ini
@@ -0,0 +1,6 @@
+# pytest.ini
+[pytest]
+minversion = 7.0
+addopts = -qq --capture=no
+testpaths =
+ src/tests
diff --git a/requirements.dev.txt b/requirements.dev.txt
new file mode 100644
index 00000000..722ab0dd
--- /dev/null
+++ b/requirements.dev.txt
@@ -0,0 +1,7 @@
+-r requirements.txt
+flake8>=6.0.0
+freezegun>=1.2.2
+pre-commit>=3.3.3
+pytest-mock>=3.11.1
+pytest>=7.4.0
+syrupy>=4.0.4
diff --git a/requirements.txt b/requirements.txt
index 505865c9..761d2e51 100644
--- a/requirements.txt
+++ b/requirements.txt
@@ -1,4 +1,8 @@
-imutils>=0.5.2
-matplotlib>=3.0.2
-numpy>=1.16.0
-pandas>=0.24.0
+deepmerge>=1.1.0
+dotmap>=1.3.30
+jsonschema>=4.17.3
+matplotlib>=3.7.1
+numpy>=1.25.0
+pandas>=2.0.2
+rich>=13.4.2
+screeninfo>=0.8.1
diff --git a/samples/answer-key/using-csv/adrian_omr.png b/samples/answer-key/using-csv/adrian_omr.png
new file mode 100644
index 00000000..d8db0994
Binary files /dev/null and b/samples/answer-key/using-csv/adrian_omr.png differ
diff --git a/samples/answer-key/using-csv/answer_key.csv b/samples/answer-key/using-csv/answer_key.csv
new file mode 100644
index 00000000..566201d1
--- /dev/null
+++ b/samples/answer-key/using-csv/answer_key.csv
@@ -0,0 +1,5 @@
+q1,C
+q2,E
+q3,A
+q4,B
+q5,B
\ No newline at end of file
diff --git a/samples/answer-key/using-csv/evaluation.json b/samples/answer-key/using-csv/evaluation.json
new file mode 100644
index 00000000..14a3db25
--- /dev/null
+++ b/samples/answer-key/using-csv/evaluation.json
@@ -0,0 +1,14 @@
+{
+ "source_type": "csv",
+ "options": {
+ "answer_key_csv_path": "answer_key.csv",
+ "should_explain_scoring": true
+ },
+ "marking_schemes": {
+ "DEFAULT": {
+ "correct": "1",
+ "incorrect": "0",
+ "unmarked": "0"
+ }
+ }
+}
diff --git a/samples/answer-key/using-csv/template.json b/samples/answer-key/using-csv/template.json
new file mode 100644
index 00000000..41ec9ffa
--- /dev/null
+++ b/samples/answer-key/using-csv/template.json
@@ -0,0 +1,35 @@
+{
+ "pageDimensions": [
+ 300,
+ 400
+ ],
+ "bubbleDimensions": [
+ 25,
+ 25
+ ],
+ "preProcessors": [
+ {
+ "name": "CropPage",
+ "options": {
+ "morphKernel": [
+ 10,
+ 10
+ ]
+ }
+ }
+ ],
+ "fieldBlocks": {
+ "MCQ_Block_1": {
+ "fieldType": "QTYPE_MCQ5",
+ "origin": [
+ 65,
+ 60
+ ],
+ "fieldLabels": [
+ "q1..5"
+ ],
+ "labelsGap": 52,
+ "bubblesGap": 41
+ }
+ }
+}
diff --git a/samples/answer-key/weighted-answers/evaluation.json b/samples/answer-key/weighted-answers/evaluation.json
new file mode 100644
index 00000000..c0daefcf
--- /dev/null
+++ b/samples/answer-key/weighted-answers/evaluation.json
@@ -0,0 +1,35 @@
+{
+ "source_type": "custom",
+ "options": {
+ "questions_in_order": [
+ "q1..5"
+ ],
+ "answers_in_order": [
+ "C",
+ "E",
+ [
+ "A",
+ "C"
+ ],
+ [
+ [
+ "B",
+ 2
+ ],
+ [
+ "C",
+ "3/2"
+ ]
+ ],
+ "C"
+ ],
+ "should_explain_scoring": true
+ },
+ "marking_schemes": {
+ "DEFAULT": {
+ "correct": "3",
+ "incorrect": "-1",
+ "unmarked": "0"
+ }
+ }
+}
diff --git a/samples/answer-key/weighted-answers/images/adrian_omr.png b/samples/answer-key/weighted-answers/images/adrian_omr.png
new file mode 100644
index 00000000..69a53823
Binary files /dev/null and b/samples/answer-key/weighted-answers/images/adrian_omr.png differ
diff --git a/samples/answer-key/weighted-answers/images/adrian_omr_2.png b/samples/answer-key/weighted-answers/images/adrian_omr_2.png
new file mode 100644
index 00000000..d8db0994
Binary files /dev/null and b/samples/answer-key/weighted-answers/images/adrian_omr_2.png differ
diff --git a/samples/answer-key/weighted-answers/template.json b/samples/answer-key/weighted-answers/template.json
new file mode 100644
index 00000000..41ec9ffa
--- /dev/null
+++ b/samples/answer-key/weighted-answers/template.json
@@ -0,0 +1,35 @@
+{
+ "pageDimensions": [
+ 300,
+ 400
+ ],
+ "bubbleDimensions": [
+ 25,
+ 25
+ ],
+ "preProcessors": [
+ {
+ "name": "CropPage",
+ "options": {
+ "morphKernel": [
+ 10,
+ 10
+ ]
+ }
+ }
+ ],
+ "fieldBlocks": {
+ "MCQ_Block_1": {
+ "fieldType": "QTYPE_MCQ5",
+ "origin": [
+ 65,
+ 60
+ ],
+ "fieldLabels": [
+ "q1..5"
+ ],
+ "labelsGap": 52,
+ "bubblesGap": 41
+ }
+ }
+}
diff --git a/samples/community/Antibodyy/simple_omr_sheet.jpg b/samples/community/Antibodyy/simple_omr_sheet.jpg
new file mode 100644
index 00000000..661d5f4f
Binary files /dev/null and b/samples/community/Antibodyy/simple_omr_sheet.jpg differ
diff --git a/samples/community/Antibodyy/template.json b/samples/community/Antibodyy/template.json
new file mode 100644
index 00000000..7e962bbf
--- /dev/null
+++ b/samples/community/Antibodyy/template.json
@@ -0,0 +1,35 @@
+{
+ "pageDimensions": [
+ 299,
+ 398
+ ],
+ "bubbleDimensions": [
+ 42,
+ 42
+ ],
+ "fieldBlocks": {
+ "MCQBlock1": {
+ "fieldType": "QTYPE_MCQ5",
+ "origin": [
+ 65,
+ 79
+ ],
+ "bubblesGap": 43,
+ "labelsGap": 50,
+ "fieldLabels": [
+ "q1..6"
+ ]
+ }
+ },
+ "preProcessors": [
+ {
+ "name": "CropPage",
+ "options": {
+ "morphKernel": [
+ 10,
+ 10
+ ]
+ }
+ }
+ ]
+}
diff --git a/samples/community/Sandeep-1507/omr-1.png b/samples/community/Sandeep-1507/omr-1.png
new file mode 100644
index 00000000..8c40655e
Binary files /dev/null and b/samples/community/Sandeep-1507/omr-1.png differ
diff --git a/samples/community/Sandeep-1507/omr-2.png b/samples/community/Sandeep-1507/omr-2.png
new file mode 100644
index 00000000..aabef0a5
Binary files /dev/null and b/samples/community/Sandeep-1507/omr-2.png differ
diff --git a/samples/community/Sandeep-1507/omr-3.png b/samples/community/Sandeep-1507/omr-3.png
new file mode 100644
index 00000000..8f1cb5c8
Binary files /dev/null and b/samples/community/Sandeep-1507/omr-3.png differ
diff --git a/samples/community/Sandeep-1507/template.json b/samples/community/Sandeep-1507/template.json
new file mode 100644
index 00000000..b35404e2
--- /dev/null
+++ b/samples/community/Sandeep-1507/template.json
@@ -0,0 +1,234 @@
+{
+ "pageDimensions": [
+ 1189,
+ 1682
+ ],
+ "bubbleDimensions": [
+ 15,
+ 15
+ ],
+ "preProcessors": [
+ {
+ "name": "GaussianBlur",
+ "options": {
+ "kSize": [
+ 3,
+ 3
+ ],
+ "sigmaX": 0
+ }
+ }
+ ],
+ "customLabels": {
+ "Booklet_No": [
+ "b1..7"
+ ]
+ },
+ "fieldBlocks": {
+ "Booklet_No": {
+ "fieldType": "QTYPE_INT",
+ "origin": [
+ 112,
+ 530
+ ],
+ "fieldLabels": [
+ "b1..7"
+ ],
+ "emptyValue": "no",
+ "bubblesGap": 28,
+ "labelsGap": 26.5
+ },
+ "MCQBlock1a1": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q1..10"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 476,
+ 100
+ ]
+ },
+ "MCQBlock1a2": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q11..20"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 476,
+ 370
+ ]
+ },
+ "MCQBlock1a3": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q21..35"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 476,
+ 638
+ ]
+ },
+ "MCQBlock2a1": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q51..60"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 645,
+ 100
+ ]
+ },
+ "MCQBlock2a2": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q61..70"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 645,
+ 370
+ ]
+ },
+ "MCQBlock2a3": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q71..85"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 645,
+ 638
+ ]
+ },
+ "MCQBlock3a1": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q101..110"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 815,
+ 100
+ ]
+ },
+ "MCQBlock3a2": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q111..120"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 815,
+ 370
+ ]
+ },
+ "MCQBlock3a3": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q121..135"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 815,
+ 638
+ ]
+ },
+ "MCQBlock4a1": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q151..160"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 983,
+ 100
+ ]
+ },
+ "MCQBlock4a2": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q161..170"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 983,
+ 370
+ ]
+ },
+ "MCQBlock4a3": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q171..185"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 983,
+ 638
+ ]
+ },
+ "MCQBlock1a": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q36..50"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 480,
+ 1061
+ ]
+ },
+ "MCQBlock2a": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q86..100"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 648,
+ 1061
+ ]
+ },
+ "MCQBlock3a": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q136..150"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 815,
+ 1061
+ ]
+ },
+ "MCQBlock4a": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q186..200"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.6,
+ "origin": [
+ 986,
+ 1061
+ ]
+ }
+ }
+}
diff --git a/samples/community/Shamanth/omr_sheet_01.png b/samples/community/Shamanth/omr_sheet_01.png
new file mode 100644
index 00000000..ead17dd1
Binary files /dev/null and b/samples/community/Shamanth/omr_sheet_01.png differ
diff --git a/samples/community/Shamanth/template.json b/samples/community/Shamanth/template.json
new file mode 100644
index 00000000..fb18d975
--- /dev/null
+++ b/samples/community/Shamanth/template.json
@@ -0,0 +1,25 @@
+{
+ "pageDimensions": [
+ 300,
+ 400
+ ],
+ "bubbleDimensions": [
+ 20,
+ 20
+ ],
+ "fieldBlocks": {
+ "MCQBlock1": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 78,
+ 41
+ ],
+ "fieldLabels": [
+ "q21..28"
+ ],
+ "bubblesGap": 56,
+ "labelsGap": 46
+ }
+ },
+ "preProcessors": []
+}
diff --git a/samples/community/UPSC-mock/answer_key.jpg b/samples/community/UPSC-mock/answer_key.jpg
new file mode 100644
index 00000000..7a229c1b
Binary files /dev/null and b/samples/community/UPSC-mock/answer_key.jpg differ
diff --git a/samples/community/UPSC-mock/config.json b/samples/community/UPSC-mock/config.json
new file mode 100644
index 00000000..0edf9d22
--- /dev/null
+++ b/samples/community/UPSC-mock/config.json
@@ -0,0 +1,11 @@
+{
+ "dimensions": {
+ "display_height": 1800,
+ "display_width": 2400,
+ "processing_height": 2400,
+ "processing_width": 1800
+ },
+ "outputs": {
+ "show_image_level": 0
+ }
+}
diff --git a/samples/community/UPSC-mock/evaluation.json b/samples/community/UPSC-mock/evaluation.json
new file mode 100644
index 00000000..42fbec8b
--- /dev/null
+++ b/samples/community/UPSC-mock/evaluation.json
@@ -0,0 +1,18 @@
+{
+ "source_type": "csv",
+ "options": {
+ "answer_key_csv_path": "answer_key.csv",
+ "answer_key_image_path": "answer_key.jpg",
+ "questions_in_order": [
+ "q1..100"
+ ],
+ "should_explain_scoring": true
+ },
+ "marking_schemes": {
+ "DEFAULT": {
+ "correct": "2",
+ "incorrect": "-2/3",
+ "unmarked": "0"
+ }
+ }
+}
diff --git a/samples/community/UPSC-mock/scan-angles/angle-1.jpg b/samples/community/UPSC-mock/scan-angles/angle-1.jpg
new file mode 100644
index 00000000..9c75d7be
Binary files /dev/null and b/samples/community/UPSC-mock/scan-angles/angle-1.jpg differ
diff --git a/samples/community/UPSC-mock/scan-angles/angle-2.jpg b/samples/community/UPSC-mock/scan-angles/angle-2.jpg
new file mode 100644
index 00000000..edab6060
Binary files /dev/null and b/samples/community/UPSC-mock/scan-angles/angle-2.jpg differ
diff --git a/samples/community/UPSC-mock/scan-angles/angle-3.jpg b/samples/community/UPSC-mock/scan-angles/angle-3.jpg
new file mode 100644
index 00000000..f1da4af9
Binary files /dev/null and b/samples/community/UPSC-mock/scan-angles/angle-3.jpg differ
diff --git a/samples/community/UPSC-mock/template.json b/samples/community/UPSC-mock/template.json
new file mode 100644
index 00000000..a1ef5c6b
--- /dev/null
+++ b/samples/community/UPSC-mock/template.json
@@ -0,0 +1,195 @@
+{
+ "pageDimensions": [
+ 1800,
+ 2400
+ ],
+ "bubbleDimensions": [
+ 30,
+ 25
+ ],
+ "customLabels": {
+ "Subject Code": [
+ "subjectCode1",
+ "subjectCode2"
+ ],
+ "Roll": [
+ "roll1..10"
+ ]
+ },
+ "fieldBlocks": {
+ "bookletNo": {
+ "origin": [
+ 595,
+ 545
+ ],
+ "bubblesGap": 68,
+ "labelsGap": 0,
+ "fieldLabels": [
+ "bookletNo"
+ ],
+ "bubbleValues": [
+ "A",
+ "B",
+ "C",
+ "D"
+ ],
+ "direction": "vertical"
+ },
+ "subjectCode": {
+ "origin": [
+ 912,
+ 512
+ ],
+ "bubblesGap": 33,
+ "labelsGap": 42.5,
+ "fieldLabels": [
+ "subjectCode1",
+ "subjectCode2"
+ ],
+ "fieldType": "QTYPE_INT"
+ },
+ "roll": {
+ "origin": [
+ 1200,
+ 510
+ ],
+ "bubblesGap": 33,
+ "labelsGap": 42.8,
+ "fieldLabels": [
+ "roll1..10"
+ ],
+ "fieldType": "QTYPE_INT"
+ },
+ "q01block": {
+ "origin": [
+ 500,
+ 927
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q1..10"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q11block": {
+ "origin": [
+ 500,
+ 1258
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q11..20"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q21block": {
+ "origin": [
+ 500,
+ 1589
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q21..30"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q31block": {
+ "origin": [
+ 495,
+ 1925
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q31..40"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q41block": {
+ "origin": [
+ 811,
+ 927
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q41..50"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q51block": {
+ "origin": [
+ 811,
+ 1258
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q51..60"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q61block": {
+ "origin": [
+ 811,
+ 1589
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q61..70"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q71block": {
+ "origin": [
+ 811,
+ 1925
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q71..80"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q81block": {
+ "origin": [
+ 1125,
+ 927
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q81..90"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q91block": {
+ "origin": [
+ 1125,
+ 1258
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q91..100"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ }
+ },
+ "preProcessors": [
+ {
+ "name": "CropPage",
+ "options": {
+ "morphKernel": [
+ 10,
+ 10
+ ]
+ }
+ }
+ ]
+}
diff --git a/samples/community/UmarFarootAPS/answer_key.csv b/samples/community/UmarFarootAPS/answer_key.csv
new file mode 100644
index 00000000..b40e8959
--- /dev/null
+++ b/samples/community/UmarFarootAPS/answer_key.csv
@@ -0,0 +1,200 @@
+q1,C
+q2,C
+q3,"D,E"
+q4,"A,AB"
+q5,"[['A', '1'], ['B', '2']]"
+q6,"['A', 'B']"
+q7,C
+q8,D
+q9,B
+q10,B
+q11,A
+q12,A
+q13,C
+q14,B
+q15,D
+q16,B
+q17,C
+q18,A
+q19,B
+q20,D
+q21,D
+q22,C
+q23,A
+q24,C
+q25,D
+q26,C
+q27,C
+q28,B
+q29,A
+q30,D
+q31,C
+q32,B
+q33,B
+q34,C
+q35,A
+q36,D
+q37,C
+q38,B
+q39,C
+q40,A
+q41,A
+q42,C
+q43,D
+q44,D
+q45,B
+q46,C
+q47,C
+q48,A
+q49,C
+q50,B
+q51,B
+q52,C
+q53,D
+q54,C
+q55,B
+q56,B
+q57,A
+q58,A
+q59,D
+q60,C
+q61,C
+q62,B
+q63,A
+q64,C
+q65,D
+q66,C
+q67,B
+q68,A
+q69,B
+q70,B
+q71,C
+q72,B
+q73,C
+q74,A
+q75,A
+q76,C
+q77,D
+q78,D
+q79,B
+q80,A
+q81,B
+q82,C
+q83,D
+q84,C
+q85,A
+q86,C
+q87,D
+q88,B
+q89,C
+q90,B
+q91,B
+q92,A
+q93,C
+q94,D
+q95,C
+q96,B
+q97,B
+q98,A
+q99,A
+q100,A
+q101,A
+q102,B
+q103,C
+q104,C
+q105,A
+q106,D
+q107,B
+q108,A
+q109,C
+q110,B
+q111,B
+q112,C
+q113,C
+q114,B
+q115,D
+q116,B
+q117,A
+q118,C
+q119,D
+q120,C
+q121,C
+q122,A
+q123,B
+q124,C
+q125,D
+q126,C
+q127,C
+q128,D
+q129,D
+q130,A
+q131,A
+q132,C
+q133,B
+q134,C
+q135,D
+q136,B
+q137,C
+q138,A
+q139,B
+q140,D
+q141,D
+q142,C
+q143,D
+q144,A
+q145,A
+q146,C
+q147,A
+q148,D
+q149,D
+q150,B
+q151,A
+q152,B
+q153,B
+q154,D
+q155,D
+q156,B
+q157,A
+q158,B
+q159,A
+q160,C
+q161,D
+q162,C
+q163,A
+q164,B
+q165,D
+q166,D
+q167,C
+q168,C
+q169,C
+q170,D
+q171,A
+q172,A
+q173,C
+q174,C
+q175,B
+q176,D
+q177,A
+q178,B
+q179,B
+q180,C
+q181,D
+q182,C
+q183,B
+q184,B
+q185,C
+q186,D
+q187,D
+q188,A
+q189,A
+q190,B
+q191,C
+q192,B
+q193,D
+q194,C
+q195,B
+q196,B
+q197,A
+q198,B
+q199,B
+q200,A
diff --git a/samples/community/UmarFarootAPS/config.json b/samples/community/UmarFarootAPS/config.json
new file mode 100644
index 00000000..fde8f5b2
--- /dev/null
+++ b/samples/community/UmarFarootAPS/config.json
@@ -0,0 +1,11 @@
+{
+ "dimensions": {
+ "display_height": 960,
+ "display_width": 1280,
+ "processing_height": 1640,
+ "processing_width": 1332
+ },
+ "outputs": {
+ "show_image_level": 0
+ }
+}
diff --git a/samples/community/UmarFarootAPS/evaluation.json b/samples/community/UmarFarootAPS/evaluation.json
new file mode 100644
index 00000000..14a3db25
--- /dev/null
+++ b/samples/community/UmarFarootAPS/evaluation.json
@@ -0,0 +1,14 @@
+{
+ "source_type": "csv",
+ "options": {
+ "answer_key_csv_path": "answer_key.csv",
+ "should_explain_scoring": true
+ },
+ "marking_schemes": {
+ "DEFAULT": {
+ "correct": "1",
+ "incorrect": "0",
+ "unmarked": "0"
+ }
+ }
+}
diff --git a/samples/sample3/omr_marker.jpg b/samples/community/UmarFarootAPS/omr_marker.jpg
similarity index 100%
rename from samples/sample3/omr_marker.jpg
rename to samples/community/UmarFarootAPS/omr_marker.jpg
diff --git a/samples/community/UmarFarootAPS/scans/scan-type-1.jpg b/samples/community/UmarFarootAPS/scans/scan-type-1.jpg
new file mode 100644
index 00000000..0932b240
Binary files /dev/null and b/samples/community/UmarFarootAPS/scans/scan-type-1.jpg differ
diff --git a/samples/community/UmarFarootAPS/scans/scan-type-2.jpg b/samples/community/UmarFarootAPS/scans/scan-type-2.jpg
new file mode 100644
index 00000000..2eef7c32
Binary files /dev/null and b/samples/community/UmarFarootAPS/scans/scan-type-2.jpg differ
diff --git a/samples/community/UmarFarootAPS/template.json b/samples/community/UmarFarootAPS/template.json
new file mode 100644
index 00000000..f096f518
--- /dev/null
+++ b/samples/community/UmarFarootAPS/template.json
@@ -0,0 +1,188 @@
+{
+ "pageDimensions": [
+ 2550,
+ 3300
+ ],
+ "bubbleDimensions": [
+ 32,
+ 32
+ ],
+ "preProcessors": [
+ {
+ "name": "CropOnMarkers",
+ "options": {
+ "relativePath": "omr_marker.jpg",
+ "sheetToMarkerWidthRatio": 17
+ }
+ }
+ ],
+ "customLabels": {
+ "Roll_no": [
+ "r1",
+ "r2",
+ "r3",
+ "r4"
+ ]
+ },
+ "fieldBlocks": {
+ "Roll_no": {
+ "fieldType": "QTYPE_INT",
+ "origin": [
+ 2169,
+ 180
+ ],
+ "fieldLabels": [
+ "r1",
+ "r2",
+ "r3",
+ "r4"
+ ],
+ "bubblesGap": 61,
+ "labelsGap": 93
+ },
+ "MCQBlock1a1": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 197,
+ 300
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q1..17"
+ ]
+ },
+ "MCQBlock1a2": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 197,
+ 1310
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q18..34"
+ ]
+ },
+ "MCQBlock1a3": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 197,
+ 2316
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q35..50"
+ ]
+ },
+ "MCQBlock1a4": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 725,
+ 300
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q51..67"
+ ]
+ },
+ "MCQBlock1a5": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 725,
+ 1310
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q68..84"
+ ]
+ },
+ "MCQBlock1a6": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 725,
+ 2316
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q85..100"
+ ]
+ },
+ "MCQBlock1a7": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 1250,
+ 300
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q101..117"
+ ]
+ },
+ "MCQBlock1a8": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 1250,
+ 1310
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q118..134"
+ ]
+ },
+ "MCQBlock1a9": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 1250,
+ 2316
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q135..150"
+ ]
+ },
+ "MCQBlock1a10": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 1770,
+ 300
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q151..167"
+ ]
+ },
+ "MCQBlock1a11": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 1770,
+ 1310
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q168..184"
+ ]
+ },
+ "MCQBlock1a12": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 1770,
+ 2316
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q185..200"
+ ]
+ }
+ }
+}
diff --git a/samples/community/dxuian/omrcollegesheet.jpg b/samples/community/dxuian/omrcollegesheet.jpg
new file mode 100644
index 00000000..b9d70c6b
Binary files /dev/null and b/samples/community/dxuian/omrcollegesheet.jpg differ
diff --git a/samples/community/dxuian/template.json b/samples/community/dxuian/template.json
new file mode 100644
index 00000000..43bf2be6
--- /dev/null
+++ b/samples/community/dxuian/template.json
@@ -0,0 +1,48 @@
+{
+ "pageDimensions": [707, 484],
+ "bubbleDimensions": [15, 10],
+ "fieldBlocks": {
+ "Column1": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [82, 35],
+ "bubblesGap": 21,
+ "labelsGap": 22.7,
+ "bubbleCount": 20,
+ "fieldLabels": ["Q1", "Q2", "Q3", "Q4", "Q5", "Q6", "Q7", "Q8", "Q9", "Q10", "Q11", "Q12", "Q13", "Q14", "Q15", "Q16", "Q17", "Q18", "Q19", "Q20"]
+ },
+ "Column2": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [205, 35],
+ "bubblesGap": 21,
+ "labelsGap": 22.7,
+ "bubbleCount": 20,
+ "fieldLabels": ["Q21", "Q22", "Q23", "Q24", "Q25", "Q26", "Q27", "Q28", "Q29", "Q30", "Q31", "Q32", "Q33", "Q34", "Q35", "Q36", "Q37", "Q38", "Q39", "Q40"]
+ },
+ "Column3": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [327, 35],
+ "bubblesGap": 21,
+ "labelsGap": 22.7,
+ "bubbleCount": 20,
+ "fieldLabels": ["Q41", "Q42", "Q43", "Q44", "Q45", "Q46", "Q47", "Q48", "Q49", "Q50", "Q51", "Q52", "Q53", "Q54", "Q55", "Q56", "Q57", "Q58", "Q59", "Q60"]
+ },
+ "Column4": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [450, 35],
+ "bubblesGap": 21,
+ "labelsGap": 22.7,
+ "bubbleCount": 20,
+ "fieldLabels": ["Q61", "Q62", "Q63", "Q64", "Q65", "Q66", "Q67", "Q68", "Q69", "Q70", "Q71", "Q72", "Q73", "Q74", "Q75", "Q76", "Q77", "Q78", "Q79", "Q80"]
+ },
+ "Column5": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [573, 35],
+ "bubblesGap": 21,
+ "labelsGap": 22.7,
+ "bubbleCount": 20,
+ "fieldLabels": ["Q81", "Q82", "Q83", "Q84", "Q85", "Q86", "Q87", "Q88", "Q89", "Q90", "Q91", "Q92", "Q93", "Q94", "Q95", "Q96", "Q97", "Q98", "Q99", "Q100"]
+ }
+ },
+
+ "emptyValue": "-"
+}
\ No newline at end of file
diff --git a/samples/community/ibrahimkilic/template.json b/samples/community/ibrahimkilic/template.json
new file mode 100644
index 00000000..f0cb9cf3
--- /dev/null
+++ b/samples/community/ibrahimkilic/template.json
@@ -0,0 +1,30 @@
+{
+ "pageDimensions": [
+ 299,
+ 328
+ ],
+ "bubbleDimensions": [
+ 20,
+ 20
+ ],
+ "emptyValue": "no",
+ "fieldBlocks": {
+ "YesNoBlock1": {
+ "direction": "horizontal",
+ "bubbleValues": [
+ "yes"
+ ],
+ "origin": [
+ 15,
+ 55
+ ],
+ "emptyValue": "no",
+ "bubblesGap": 48,
+ "labelsGap": 48,
+ "fieldLabels": [
+ "q1..5"
+ ]
+ }
+ },
+ "preProcessors": []
+}
diff --git a/samples/community/ibrahimkilic/yes_no_questionnarie.jpg b/samples/community/ibrahimkilic/yes_no_questionnarie.jpg
new file mode 100644
index 00000000..e9436f41
Binary files /dev/null and b/samples/community/ibrahimkilic/yes_no_questionnarie.jpg differ
diff --git a/samples/community/samuelIkoli/template.json b/samples/community/samuelIkoli/template.json
new file mode 100644
index 00000000..759f6972
--- /dev/null
+++ b/samples/community/samuelIkoli/template.json
@@ -0,0 +1,28 @@
+{
+ "pageDimensions": [630, 404],
+ "bubbleDimensions": [20, 15],
+ "customLabels": {},
+ "fieldBlocks": {
+ "MCQBlock1": {
+ "fieldType": "QTYPE_MCQ5",
+ "origin": [33, 6],
+ "fieldLabels": ["q1", "q2", "q3", "q4", "q5", "q6", "q7", "q8", "q9", "q10", "q11", "q12", "q13", "q14", "q15", "q16", "q17", "q18", "q19", "q20"],
+ "bubblesGap": 33,
+ "labelsGap": 20
+ },
+ "MCQBlock2": {
+ "fieldType": "QTYPE_MCQ5",
+ "origin": [248, 6],
+ "fieldLabels": ["q21", "q22", "q23", "q24", "q25", "q26", "q27", "q28", "q29", "q30", "q31", "q32", "q33", "q34", "q35", "q36", "q37", "q38", "q39", "q40"],
+ "bubblesGap": 33,
+ "labelsGap": 20
+ },
+ "MCQBlock3": {
+ "fieldType": "QTYPE_MCQ5",
+ "origin": [465, 6],
+ "fieldLabels": ["q41", "q42", "q43", "q44", "q45", "q46", "q47", "q48", "q49", "q50", "q51", "q52", "q53", "q54", "q55", "q56", "q57", "q58", "q59", "q60"],
+ "bubblesGap": 33,
+ "labelsGap": 20
+ }
+ }
+}
diff --git a/samples/community/samuelIkoli/waec_sample.jpeg b/samples/community/samuelIkoli/waec_sample.jpeg
new file mode 100644
index 00000000..fcd61e80
Binary files /dev/null and b/samples/community/samuelIkoli/waec_sample.jpeg differ
diff --git a/samples/sample1/config.json b/samples/sample1/config.json
new file mode 100644
index 00000000..a9daf449
--- /dev/null
+++ b/samples/sample1/config.json
@@ -0,0 +1,11 @@
+{
+ "dimensions": {
+ "display_height": 2480,
+ "display_width": 1640,
+ "processing_height": 820,
+ "processing_width": 666
+ },
+ "outputs": {
+ "show_image_level": 5
+ }
+}
diff --git a/samples/sample1/template.json b/samples/sample1/template.json
index ec0f9c68..46635774 100644
--- a/samples/sample1/template.json
+++ b/samples/sample1/template.json
@@ -1,260 +1,197 @@
{
- "Dimensions": [
+ "pageDimensions": [
1846,
1500
],
- "BubbleDimensions": [
+ "bubbleDimensions": [
40,
40
],
- "Options": {
- "Marker": {
- "RelativePath": "omr_marker.jpg",
- "SheetToMarkerWidthRatio": 17
- }
- },
- "Concatenations": {
+ "customLabels": {
"Roll": [
- "Squad",
"Medium",
- "roll0",
- "roll1",
- "roll2",
- "roll3",
- "roll4",
- "roll5",
- "roll6",
- "roll7",
- "roll8"
+ "roll1..9"
],
"q5": [
- "q5.1",
- "q5.2"
+ "q5_1",
+ "q5_2"
],
"q6": [
- "q6.1",
- "q6.2"
+ "q6_1",
+ "q6_2"
],
"q7": [
- "q7.1",
- "q7.2"
+ "q7_1",
+ "q7_2"
],
- "q9": [
- "q9.1",
- "q9.2"
+ "q8": [
+ "q8_1",
+ "q8_2"
],
- "q11": [
- "q8.1",
- "q8.2"
+ "q9": [
+ "q9_1",
+ "q9_2"
]
},
- "Singles": [
- "q1",
- "q2",
- "q3",
- "q4",
- "q10",
- "q11",
- "q12",
- "q13",
- "q14",
- "q15",
- "q16",
- "q17",
- "q18",
- "q19",
- "q20"
- ],
- "QBlocks": {
+ "fieldBlocks": {
"Medium": {
- "qType": "QTYPE_MED",
- "orig": [
- 160,
- 285
- ],
- "bigGaps": [
- 115,
- 11
- ],
- "gaps": [
- 59,
- 46
- ],
- "qNos": [
- [
- [
- "Medium"
- ]
- ]
+ "bubblesGap": 41,
+ "bubbleValues": [
+ "E",
+ "H"
+ ],
+ "direction": "vertical",
+ "fieldLabels": [
+ "Medium"
+ ],
+ "labelsGap": 0,
+ "origin": [
+ 170,
+ 282
]
},
"Roll": {
- "qType": "QTYPE_ROLL",
- "orig": [
- 218,
- 285
- ],
- "bigGaps": [
- 115,
- 11
- ],
- "gaps": [
- 58,
- 46
- ],
- "qNos": [
- [
- [
- "roll0",
- "roll1",
- "roll2",
- "roll3",
- "roll4",
- "roll5",
- "roll6",
- "roll7",
- "roll8"
- ]
- ]
+ "fieldType": "QTYPE_INT",
+ "fieldLabels": [
+ "roll1..9"
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 58,
+ "origin": [
+ 225,
+ 282
]
},
- "Int1": {
- "qType": "QTYPE_INT",
- "orig": [
+ "Int_Block_Q5": {
+ "fieldType": "QTYPE_INT",
+ "fieldLabels": [
+ "q5_1",
+ "q5_2"
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 60,
+ "origin": [
903,
- 285
- ],
- "bigGaps": [
- 128,
- 11
- ],
- "gaps": [
- 59,
- 46
- ],
- "qNos": [
- [
- [
- "q5.1",
- "q5.2"
- ],
- [
- "q6.1",
- "q6.2"
- ],
- [
- "q7.1",
- "q7.2"
- ]
- ]
+ 282
]
},
- "Int2": {
- "qType": "QTYPE_INT",
- "orig": [
- 1418,
- 285
- ],
- "bigGaps": [
- 128,
- 11
- ],
- "gaps": [
- 59,
- 46
- ],
- "qNos": [
- [
- [
- "q8.1",
- "q8.2"
- ],
- [
- "q9.1",
- "q9.2"
- ]
- ]
+ "Int_Block_Q6": {
+ "fieldType": "QTYPE_INT",
+ "fieldLabels": [
+ "q6_1",
+ "q6_2"
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 60,
+ "origin": [
+ 1077,
+ 282
]
},
- "Mcq1": {
- "qType": "QTYPE_MCQ4",
- "orig": [
- 118,
- 857
- ],
- "bigGaps": [
- 115,
- 181
- ],
- "gaps": [
- 59,
- 53
- ],
- "qNos": [
- [
- [
- "q1",
- "q2",
- "q3",
- "q4"
- ],
- [
- "q10",
- "q11",
- "q12",
- "q13"
- ]
- ]
+ "Int_Block_Q7": {
+ "fieldType": "QTYPE_INT",
+ "fieldLabels": [
+ "q7_1",
+ "q7_2"
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 60,
+ "origin": [
+ 1240,
+ 282
+ ]
+ },
+ "Int_Block_Q8": {
+ "fieldType": "QTYPE_INT",
+ "fieldLabels": [
+ "q8_1",
+ "q8_2"
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 57,
+ "origin": [
+ 1410,
+ 282
+ ]
+ },
+ "Int_Block_Q9": {
+ "fieldType": "QTYPE_INT",
+ "fieldLabels": [
+ "q9_1",
+ "q9_2"
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 57,
+ "origin": [
+ 1580,
+ 282
]
},
- "Mcq2": {
- "qType": "QTYPE_MCQ4",
- "orig": [
+ "MCQ_Block_Q1": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q1..4"
+ ],
+ "bubblesGap": 59,
+ "labelsGap": 50,
+ "origin": [
+ 121,
+ 860
+ ]
+ },
+ "MCQ_Block_Q10": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q10..13"
+ ],
+ "bubblesGap": 59,
+ "labelsGap": 50,
+ "origin": [
+ 121,
+ 1195
+ ]
+ },
+ "MCQ_Block_Q14": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q14..16"
+ ],
+ "bubblesGap": 57,
+ "labelsGap": 50,
+ "origin": [
905,
860
- ],
- "bigGaps": [
- 115,
- 180
- ],
- "gaps": [
- 59,
- 53
- ],
- "qNos": [
- [
- [
- "q14",
- "q15",
- "q16"
- ]
- ]
]
},
- "Mcq3": {
- "qType": "QTYPE_MCQ4",
- "orig": [
+ "MCQ_Block_Q17": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q17..20"
+ ],
+ "bubblesGap": 57,
+ "labelsGap": 50,
+ "origin": [
905,
- 1198
- ],
- "bigGaps": [
- 115,
- 180
- ],
- "gaps": [
- 59,
- 53
- ],
- "qNos": [
- [
- [
- "q17",
- "q18",
- "q19",
- "q20"
- ]
- ]
+ 1195
]
}
- }
-}
\ No newline at end of file
+ },
+ "preProcessors": [
+ {
+ "name": "CropPage",
+ "options": {
+ "morphKernel": [
+ 10,
+ 10
+ ]
+ }
+ },
+ {
+ "name": "CropOnMarkers",
+ "options": {
+ "relativePath": "omr_marker.jpg",
+ "sheetToMarkerWidthRatio": 17
+ }
+ }
+ ]
+}
diff --git a/samples/sample2/AdrianSample/adrian_omr.png b/samples/sample2/AdrianSample/adrian_omr.png
index dc008145..69a53823 100644
Binary files a/samples/sample2/AdrianSample/adrian_omr.png and b/samples/sample2/AdrianSample/adrian_omr.png differ
diff --git a/samples/sample2/AdrianSample/adrian_omr_2.png b/samples/sample2/AdrianSample/adrian_omr_2.png
new file mode 100644
index 00000000..d8db0994
Binary files /dev/null and b/samples/sample2/AdrianSample/adrian_omr_2.png differ
diff --git a/samples/sample2/config.json b/samples/sample2/config.json
new file mode 100644
index 00000000..a9daf449
--- /dev/null
+++ b/samples/sample2/config.json
@@ -0,0 +1,11 @@
+{
+ "dimensions": {
+ "display_height": 2480,
+ "display_width": 1640,
+ "processing_height": 820,
+ "processing_width": 666
+ },
+ "outputs": {
+ "show_image_level": 5
+ }
+}
diff --git a/samples/sample2/template.json b/samples/sample2/template.json
index e82a9051..41ec9ffa 100644
--- a/samples/sample2/template.json
+++ b/samples/sample2/template.json
@@ -1,46 +1,35 @@
{
- "Dimensions": [
+ "pageDimensions": [
300,
400
],
- "BubbleDimensions": [
+ "bubbleDimensions": [
25,
25
],
- "Concatenations": {},
- "Singles": [
- "q1",
- "q2",
- "q3",
- "q4",
- "q5"
+ "preProcessors": [
+ {
+ "name": "CropPage",
+ "options": {
+ "morphKernel": [
+ 10,
+ 10
+ ]
+ }
+ }
],
- "QBlocks": {
- "MCQBlock1": {
- "qType": "QTYPE_MCQ5",
- "orig": [
+ "fieldBlocks": {
+ "MCQ_Block_1": {
+ "fieldType": "QTYPE_MCQ5",
+ "origin": [
65,
60
],
- "qNos": [
- [
- [
- "q1",
- "q2",
- "q3",
- "q4",
- "q5"
- ]
- ]
- ],
- "gaps": [
- 41,
- 52
+ "fieldLabels": [
+ "q1..5"
],
- "bigGaps": [
- 30,
- 30
- ]
+ "labelsGap": 52,
+ "bubblesGap": 41
}
}
-}
\ No newline at end of file
+}
diff --git a/samples/sample3/README.md b/samples/sample3/README.md
new file mode 100644
index 00000000..d764c8b3
--- /dev/null
+++ b/samples/sample3/README.md
@@ -0,0 +1,31 @@
+## Observation
+The OMR layout is slightly different on colored thick papers vs on xeroxed thin papers. The shift becomes noticible in case of OMR with large number of questions.
+
+We overlapped a colored OMR sheet with a xerox copy of the same OMR sheet(both printed on A4 papers) and observed that there is a great amount of layout sheet as we reach the bottom of the OMR.
+
+Link to an explainer with a real life example: [Google Drive](https://drive.google.com/drive/folders/1GpZTmpEhEjSALJEMjHwDafzWKgEoCTOI?usp=sharing)
+
+## Reasons for shifts in Template layout:
+Listing out a few reasons for the above observation:
+### Printer margin setting
+The margin settings for different printers may be different for the same OMR layout. Thus causing the print to become elongated either horizontally, vertically or both ways.
+
+### The Fan-out effect
+The fan-out effect is usually observed in a sheet fed offset press. Depending on how the papers are made, their dimensions have a tendency to change when they are exposed to moisture or water.
+
+The standard office papers(80 gsm) can easily capture moisture and change shape e.g. in case they get stored for multiple days in a place where the weather is highly humid.
+
+Below are some examples of the GSM ranges:
+
+- 74gsm to 90gsm – This is the basic standard office paper, used in your laser printers.
+- 100gsm to 120gsm – This is stationary paper used for standard letterheads, complimentary slips.
+- 130 to 170gsm – Mostly used for leaflets, posters, single-sided flyers, and brochures.
+
+## Solution
+
+It is recommended to scan each types of prints into different folders and use a separate template.json layout for each of the folders. The same is presented in this sample folder.
+
+## References
+
+- [Paper dimensional stability in sheet-fed offset printing](https://www.diva-portal.org/smash/get/diva2:517895/FULLTEXT01.pdf)
+- An analysis of a few ["Interesting" bubble sheets](https://imgur.com/a/10qwL)
diff --git a/samples/sample3/colored-thick-sheet/rgb-100-gsm.jpg b/samples/sample3/colored-thick-sheet/rgb-100-gsm.jpg
new file mode 100644
index 00000000..4a34731a
Binary files /dev/null and b/samples/sample3/colored-thick-sheet/rgb-100-gsm.jpg differ
diff --git a/samples/sample3/colored-thick-sheet/template.json b/samples/sample3/colored-thick-sheet/template.json
new file mode 100644
index 00000000..743723d6
--- /dev/null
+++ b/samples/sample3/colored-thick-sheet/template.json
@@ -0,0 +1,143 @@
+{
+ "pageDimensions": [
+ 1800,
+ 2400
+ ],
+ "bubbleDimensions": [
+ 23,
+ 20
+ ],
+ "fieldBlocks": {
+ "q01block": {
+ "origin": [
+ 504,
+ 927
+ ],
+ "bubblesGap": 60.35,
+ "labelsGap": 31.75,
+ "fieldLabels": [
+ "q1..10"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q11block": {
+ "origin": [
+ 504,
+ 1242
+ ],
+ "bubblesGap": 60.35,
+ "labelsGap": 31.75,
+ "fieldLabels": [
+ "q11..20"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q21block": {
+ "origin": [
+ 500,
+ 1562
+ ],
+ "bubblesGap": 61.25,
+ "labelsGap": 32.5,
+ "fieldLabels": [
+ "q21..30"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q31block": {
+ "origin": [
+ 500,
+ 1885
+ ],
+ "bubblesGap": 62.25,
+ "labelsGap": 33.5,
+ "fieldLabels": [
+ "q31..40"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q41block": {
+ "origin": [
+ 811,
+ 927
+ ],
+ "bubblesGap": 60.35,
+ "labelsGap": 31.75,
+ "fieldLabels": [
+ "q41..50"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q51block": {
+ "origin": [
+ 811,
+ 1242
+ ],
+ "bubblesGap": 60.35,
+ "labelsGap": 31.75,
+ "fieldLabels": [
+ "q51..60"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q61block": {
+ "origin": [
+ 811,
+ 1562
+ ],
+ "bubblesGap": 61.25,
+ "labelsGap": 32.5,
+ "fieldLabels": [
+ "q61..70"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q71block": {
+ "origin": [
+ 811,
+ 1885
+ ],
+ "bubblesGap": 62.25,
+ "labelsGap": 33.5,
+ "fieldLabels": [
+ "q71..80"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q81block": {
+ "origin": [
+ 1120,
+ 927
+ ],
+ "bubblesGap": 60.35,
+ "labelsGap": 31.75,
+ "fieldLabels": [
+ "q81..90"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q91block": {
+ "origin": [
+ 1120,
+ 1242
+ ],
+ "bubblesGap": 60.35,
+ "labelsGap": 31.75,
+ "fieldLabels": [
+ "q91..100"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ }
+ },
+ "preProcessors": [
+ {
+ "name": "CropPage",
+ "options": {
+ "morphKernel": [
+ 10,
+ 10
+ ]
+ }
+ }
+ ]
+}
diff --git a/samples/sample3/config.json b/samples/sample3/config.json
new file mode 100644
index 00000000..a9daf449
--- /dev/null
+++ b/samples/sample3/config.json
@@ -0,0 +1,11 @@
+{
+ "dimensions": {
+ "display_height": 2480,
+ "display_width": 1640,
+ "processing_height": 820,
+ "processing_width": 666
+ },
+ "outputs": {
+ "show_image_level": 5
+ }
+}
diff --git a/samples/sample3/template.json b/samples/sample3/template.json
deleted file mode 100644
index 2dddf37e..00000000
--- a/samples/sample3/template.json
+++ /dev/null
@@ -1,248 +0,0 @@
-{
- "Dimensions": [
- 1846,
- 1500
- ],
- "BubbleDimensions": [
- 40,
- 40
- ],
- "Options": {
- "Marker": {
- "RelativePath": "omr_marker.jpg",
- "SheetToMarkerWidthRatio": 17
- },
- "OverrideFlags":{
- "noCropping": true
- }
- },
- "Concatenations": {
- "Roll": [
- "Squad",
- "Medium",
- "roll0",
- "roll1",
- "roll2",
- "roll3",
- "roll4",
- "roll5",
- "roll6",
- "roll7",
- "roll8"
- ],
- "q6": [
- "q6.1",
- "q6.2"
- ],
- "q7": [
- "q7.1",
- "q7.2"
- ],
- "q8": [
- "q8.1",
- "q8.2"
- ],
- "q9": [
- "q9.1",
- "q9.2"
- ],
- "q10": [
- "q10.1",
- "q10.2"
- ]
- },
- "Singles": [
- "q1",
- "q2",
- "q3",
- "q4",
- "q5",
- "q11",
- "q12",
- "q13",
- "q14",
- "q15",
- "q16",
- "q17",
- "q18",
- "q19",
- "q20",
- "q21",
- "q22"
- ],
- "QBlocks": {
- "Medium": {
- "qType": "QTYPE_MED",
- "orig": [
- 208,
- 205
- ],
- "bigGaps": [
- 115,
- 11
- ],
- "gaps": [
- 59,
- 46
- ],
- "qNos": [
- [
- [
- "Medium"
- ]
- ]
- ]
- },
- "Roll": {
- "qType": "QTYPE_ROLL",
- "orig": [
- 261,
- 210
- ],
- "bigGaps": [
- 115,
- 11
- ],
- "gaps": [
- 58,
- 46
- ],
- "qNos": [
- [
- [
- "roll0",
- "roll1",
- "roll2",
- "roll3",
- "roll4",
- "roll5",
- "roll6",
- "roll7",
- "roll8"
- ]
- ]
- ]
- },
- "Int1": {
- "qType": "QTYPE_INT",
- "orig": [
- 935,
- 211
- ],
- "bigGaps": [
- 124,
- 11
- ],
- "gaps": [
- 57,
- 46
- ],
- "qNos": [
- [
- [
- "q6.1",
- "q6.2"
- ],
- [
- "q7.1",
- "q7.2"
- ],
- [
- "q8.1",
- "q8.2"
- ],
- [
- "q9.1",
- "q9.2"
- ],
- [
- "q10.1",
- "q10.2"
- ]
- ]
- ]
- },
- "Mcq1": {
- "qType": "QTYPE_MCQ4",
- "orig": [
- 198,
- 826
- ],
- "bigGaps": [
- 115,
- 183
- ],
- "gaps": [
- 93,
- 62
- ],
- "qNos": [
- [
- [
- "q1",
- "q2",
- "q3",
- "q4",
- "q5"
- ]
- ]
- ]
- },
- "Mcq2": {
- "qType": "QTYPE_MCQ4",
- "orig": [
- 833,
- 830
- ],
- "bigGaps": [
- 127,
- 254
- ],
- "gaps": [
- 71,
- 61
- ],
- "qNos": [
- [
- [
- "q11",
- "q12",
- "q13",
- "q14"
- ],
- [
- "q15",
- "q16",
- "q17",
- "q18"
- ]
- ]
- ]
- },
- "Mcq3": {
- "qType": "QTYPE_MCQ4",
- "orig": [
- 1481,
- 830
- ],
- "bigGaps": [
- 115,
- 183
- ],
- "gaps": [
- 73,
- 61
- ],
- "qNos": [
- [
- [
- "q19",
- "q20",
- "q21",
- "q22"
- ]
- ]
- ]
- }
- }
-}
\ No newline at end of file
diff --git a/samples/sample3/xeroxed-thin-sheet/grayscale-80-gsm.jpg b/samples/sample3/xeroxed-thin-sheet/grayscale-80-gsm.jpg
new file mode 100644
index 00000000..3e0e3efa
Binary files /dev/null and b/samples/sample3/xeroxed-thin-sheet/grayscale-80-gsm.jpg differ
diff --git a/samples/sample3/xeroxed-thin-sheet/template.json b/samples/sample3/xeroxed-thin-sheet/template.json
new file mode 100644
index 00000000..8aa4e41c
--- /dev/null
+++ b/samples/sample3/xeroxed-thin-sheet/template.json
@@ -0,0 +1,143 @@
+{
+ "pageDimensions": [
+ 1800,
+ 2400
+ ],
+ "bubbleDimensions": [
+ 23,
+ 20
+ ],
+ "fieldBlocks": {
+ "q01block": {
+ "origin": [
+ 492,
+ 924
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q1..10"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q11block": {
+ "origin": [
+ 492,
+ 1258
+ ],
+ "bubblesGap": 59.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q11..20"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q21block": {
+ "origin": [
+ 492,
+ 1589
+ ],
+ "bubblesGap": 60.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q21..30"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q31block": {
+ "origin": [
+ 487,
+ 1920
+ ],
+ "bubblesGap": 61.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q31..40"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q41block": {
+ "origin": [
+ 807,
+ 924
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q41..50"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q51block": {
+ "origin": [
+ 803,
+ 1258
+ ],
+ "bubblesGap": 59.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q51..60"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q61block": {
+ "origin": [
+ 803,
+ 1589
+ ],
+ "bubblesGap": 60.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q61..70"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q71block": {
+ "origin": [
+ 803,
+ 1920
+ ],
+ "bubblesGap": 60.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q71..80"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q81block": {
+ "origin": [
+ 1115,
+ 924
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q81..90"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q91block": {
+ "origin": [
+ 1115,
+ 1258
+ ],
+ "bubblesGap": 59.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q91..100"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ }
+ },
+ "preProcessors": [
+ {
+ "name": "CropPage",
+ "options": {
+ "morphKernel": [
+ 10,
+ 10
+ ]
+ }
+ }
+ ]
+}
diff --git a/samples/sample4/Class1/sheet1.jpg b/samples/sample4/Class1/sheet1.jpg
deleted file mode 100644
index f3248903..00000000
Binary files a/samples/sample4/Class1/sheet1.jpg and /dev/null differ
diff --git a/samples/sample4/Class2/sheet1.jpg b/samples/sample4/Class2/sheet1.jpg
deleted file mode 100644
index f3248903..00000000
Binary files a/samples/sample4/Class2/sheet1.jpg and /dev/null differ
diff --git a/samples/sample4/IMG_20201116_143512.jpg b/samples/sample4/IMG_20201116_143512.jpg
new file mode 100644
index 00000000..2f591c7a
Binary files /dev/null and b/samples/sample4/IMG_20201116_143512.jpg differ
diff --git a/samples/sample4/IMG_20201116_150717658.jpg b/samples/sample4/IMG_20201116_150717658.jpg
new file mode 100644
index 00000000..592fe425
Binary files /dev/null and b/samples/sample4/IMG_20201116_150717658.jpg differ
diff --git a/samples/sample4/IMG_20201116_150750830.jpg b/samples/sample4/IMG_20201116_150750830.jpg
new file mode 100644
index 00000000..6846ef31
Binary files /dev/null and b/samples/sample4/IMG_20201116_150750830.jpg differ
diff --git a/samples/sample4/config.json b/samples/sample4/config.json
new file mode 100644
index 00000000..df59a460
--- /dev/null
+++ b/samples/sample4/config.json
@@ -0,0 +1,13 @@
+{
+ "dimensions": {
+ "display_width": 1189,
+ "display_height": 1682
+ },
+ "threshold_params": {
+ "MIN_JUMP": 30
+ },
+ "outputs": {
+ "filter_out_multimarked_files": false,
+ "show_image_level": 5
+ }
+}
diff --git a/samples/sample4/evaluation.json b/samples/sample4/evaluation.json
new file mode 100644
index 00000000..50db0fbe
--- /dev/null
+++ b/samples/sample4/evaluation.json
@@ -0,0 +1,34 @@
+{
+ "source_type": "custom",
+ "options": {
+ "questions_in_order": [
+ "q1..11"
+ ],
+ "answers_in_order": [
+ "B",
+ "D",
+ "C",
+ "B",
+ "D",
+ "C",
+ [
+ "B",
+ "C",
+ "BC"
+ ],
+ "A",
+ "C",
+ "D",
+ "C"
+ ],
+ "should_explain_scoring": true,
+ "enable_evaluation_table_to_csv": true
+ },
+ "marking_schemes": {
+ "DEFAULT": {
+ "correct": "3",
+ "incorrect": "-1",
+ "unmarked": "0"
+ }
+ }
+}
diff --git a/samples/sample4/omr_marker.jpg b/samples/sample4/omr_marker.jpg
deleted file mode 100644
index 201dab0a..00000000
Binary files a/samples/sample4/omr_marker.jpg and /dev/null differ
diff --git a/samples/sample4/template.json b/samples/sample4/template.json
index 958d6ef8..0d615b87 100644
--- a/samples/sample4/template.json
+++ b/samples/sample4/template.json
@@ -1,263 +1,45 @@
{
- "Dimensions": [
- 1846,
- 1500
+ "pageDimensions": [
+ 1189,
+ 1682
],
- "BubbleDimensions": [
- 40,
- 40
+ "bubbleDimensions": [
+ 30,
+ 30
],
- "Options": {
- "Marker": {
- "RelativePath": "omr_marker.jpg",
- "SheetToMarkerWidthRatio": 17
+ "preProcessors": [
+ {
+ "name": "GaussianBlur",
+ "options": {
+ "kSize": [
+ 3,
+ 3
+ ],
+ "sigmaX": 0
+ }
},
- "OverrideFlags": {
- "noCropping": true
+ {
+ "name": "CropPage",
+ "options": {
+ "morphKernel": [
+ 10,
+ 10
+ ]
+ }
}
- },
- "Concatenations": {
- "Roll": [
- "Squad",
- "Medium",
- "roll0",
- "roll1",
- "roll2",
- "roll3",
- "roll4",
- "roll5",
- "roll6",
- "roll7",
- "roll8"
- ],
- "q5": [
- "q5.1",
- "q5.2"
- ],
- "q6": [
- "q6.1",
- "q6.2"
- ],
- "q7": [
- "q7.1",
- "q7.2"
- ],
- "q9": [
- "q9.1",
- "q9.2"
- ],
- "q11": [
- "q8.1",
- "q8.2"
- ]
- },
- "Singles": [
- "q1",
- "q2",
- "q3",
- "q4",
- "q10",
- "q11",
- "q12",
- "q13",
- "q14",
- "q15",
- "q16",
- "q17",
- "q18",
- "q19",
- "q20"
],
- "QBlocks": {
- "Medium": {
- "qType": "QTYPE_MED",
- "orig": [
- 160,
- 285
- ],
- "bigGaps": [
- 115,
- 11
- ],
- "gaps": [
- 59,
- 46
- ],
- "qNos": [
- [
- [
- "Medium"
- ]
- ]
- ]
- },
- "Roll": {
- "qType": "QTYPE_ROLL",
- "orig": [
- 218,
- 285
- ],
- "bigGaps": [
- 115,
- 11
- ],
- "gaps": [
- 58,
- 46
- ],
- "qNos": [
- [
- [
- "roll0",
- "roll1",
- "roll2",
- "roll3",
- "roll4",
- "roll5",
- "roll6",
- "roll7",
- "roll8"
- ]
- ]
- ]
- },
- "Int1": {
- "qType": "QTYPE_INT",
- "orig": [
- 903,
- 285
- ],
- "bigGaps": [
- 128,
- 11
- ],
- "gaps": [
- 59,
- 46
- ],
- "qNos": [
- [
- [
- "q5.1",
- "q5.2"
- ],
- [
- "q6.1",
- "q6.2"
- ],
- [
- "q7.1",
- "q7.2"
- ]
- ]
- ]
- },
- "Int2": {
- "qType": "QTYPE_INT",
- "orig": [
- 1418,
- 285
- ],
- "bigGaps": [
- 128,
- 11
- ],
- "gaps": [
- 59,
- 46
- ],
- "qNos": [
- [
- [
- "q8.1",
- "q8.2"
- ],
- [
- "q9.1",
- "q9.2"
- ]
- ]
- ]
- },
- "Mcq1": {
- "qType": "QTYPE_MCQ4",
- "orig": [
- 118,
- 857
- ],
- "bigGaps": [
- 115,
- 181
- ],
- "gaps": [
- 59,
- 53
- ],
- "qNos": [
- [
- [
- "q1",
- "q2",
- "q3",
- "q4"
- ],
- [
- "q10",
- "q11",
- "q12",
- "q13"
- ]
- ]
- ]
- },
- "Mcq2": {
- "qType": "QTYPE_MCQ4",
- "orig": [
- 905,
- 860
- ],
- "bigGaps": [
- 115,
- 180
- ],
- "gaps": [
- 59,
- 53
- ],
- "qNos": [
- [
- [
- "q14",
- "q15",
- "q16"
- ]
- ]
- ]
- },
- "Mcq3": {
- "qType": "QTYPE_MCQ4",
- "orig": [
- 905,
- 1198
- ],
- "bigGaps": [
- 115,
- 180
- ],
- "gaps": [
- 59,
- 53
- ],
- "qNos": [
- [
- [
- "q17",
- "q18",
- "q19",
- "q20"
- ]
- ]
- ]
+ "fieldBlocks": {
+ "MCQBlock1": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 134,
+ 684
+ ],
+ "fieldLabels": [
+ "q1..11"
+ ],
+ "bubblesGap": 79,
+ "labelsGap": 62
}
}
-}
\ No newline at end of file
+}
diff --git a/samples/sample5/README.md b/samples/sample5/README.md
new file mode 100644
index 00000000..b5ed8ccb
--- /dev/null
+++ b/samples/sample5/README.md
@@ -0,0 +1,6 @@
+### OMRChecker Sample
+
+This sample demonstrates multiple things, namely -
+- Running OMRChecker on images scanned using popular document scanning apps
+- Using a common template.json file for sub-folders (e.g. multiple scan batches)
+- Using evaluation.json file with custom marking (without streak-based marking)
diff --git a/samples/sample3/CamScanner/sheet2.jpg b/samples/sample5/ScanBatch1/camscanner-1.jpg
similarity index 100%
rename from samples/sample3/CamScanner/sheet2.jpg
rename to samples/sample5/ScanBatch1/camscanner-1.jpg
diff --git a/samples/sample3/CamScanner/sheet1.jpg b/samples/sample5/ScanBatch2/camscanner-2.jpg
similarity index 100%
rename from samples/sample3/CamScanner/sheet1.jpg
rename to samples/sample5/ScanBatch2/camscanner-2.jpg
diff --git a/samples/sample5/config.json b/samples/sample5/config.json
new file mode 100644
index 00000000..a9daf449
--- /dev/null
+++ b/samples/sample5/config.json
@@ -0,0 +1,11 @@
+{
+ "dimensions": {
+ "display_height": 2480,
+ "display_width": 1640,
+ "processing_height": 820,
+ "processing_width": 666
+ },
+ "outputs": {
+ "show_image_level": 5
+ }
+}
diff --git a/samples/sample5/evaluation.json b/samples/sample5/evaluation.json
new file mode 100644
index 00000000..3332fe6a
--- /dev/null
+++ b/samples/sample5/evaluation.json
@@ -0,0 +1,93 @@
+{
+ "source_type": "custom",
+ "options": {
+ "questions_in_order": [
+ "q1..22"
+ ],
+ "answers_in_order": [
+ "C",
+ "C",
+ "B",
+ "C",
+ "C",
+ [
+ "1",
+ "01"
+ ],
+ "19",
+ "10",
+ "10",
+ "18",
+ "D",
+ "A",
+ "D",
+ "D",
+ "D",
+ "C",
+ "C",
+ "C",
+ "C",
+ "D",
+ "B",
+ "A"
+ ],
+ "should_explain_scoring": true
+ },
+ "marking_schemes": {
+ "DEFAULT": {
+ "correct": "1",
+ "incorrect": "0",
+ "unmarked": "0"
+ },
+ "BOOMERANG_1": {
+ "questions": [
+ "q1..5"
+ ],
+ "marking": {
+ "correct": 4,
+ "incorrect": -1,
+ "unmarked": 0
+ }
+ },
+ "PROXIMITY_1": {
+ "questions": [
+ "q6..10"
+ ],
+ "marking": {
+ "correct": 3,
+ "incorrect": -1,
+ "unmarked": 0
+ }
+ },
+ "FIBONACCI_SECTION_1": {
+ "questions": [
+ "q11..14"
+ ],
+ "marking": {
+ "correct": 2,
+ "incorrect": -1,
+ "unmarked": 0
+ }
+ },
+ "POWER_SECTION_1": {
+ "questions": [
+ "q15..18"
+ ],
+ "marking": {
+ "correct": 1,
+ "incorrect": 0,
+ "unmarked": 0
+ }
+ },
+ "FIBONACCI_SECTION_2": {
+ "questions": [
+ "q19..22"
+ ],
+ "marking": {
+ "correct": 2,
+ "incorrect": -1,
+ "unmarked": 0
+ }
+ }
+ }
+}
diff --git a/samples/sample5/omr_marker.jpg b/samples/sample5/omr_marker.jpg
new file mode 100644
index 00000000..0929feec
Binary files /dev/null and b/samples/sample5/omr_marker.jpg differ
diff --git a/samples/sample5/template.json b/samples/sample5/template.json
new file mode 100644
index 00000000..39f6e83d
--- /dev/null
+++ b/samples/sample5/template.json
@@ -0,0 +1,188 @@
+{
+ "pageDimensions": [
+ 1846,
+ 1500
+ ],
+ "bubbleDimensions": [
+ 40,
+ 40
+ ],
+ "preProcessors": [
+ {
+ "name": "CropOnMarkers",
+ "options": {
+ "relativePath": "omr_marker.jpg",
+ "sheetToMarkerWidthRatio": 17
+ }
+ }
+ ],
+ "customLabels": {
+ "Roll": [
+ "Medium",
+ "roll1..9"
+ ],
+ "q6": [
+ "q6_1",
+ "q6_2"
+ ],
+ "q7": [
+ "q7_1",
+ "q7_2"
+ ],
+ "q8": [
+ "q8_1",
+ "q8_2"
+ ],
+ "q9": [
+ "q9_1",
+ "q9_2"
+ ],
+ "q10": [
+ "q10_1",
+ "q10_2"
+ ]
+ },
+ "fieldBlocks": {
+ "Medium": {
+ "bubbleValues": [
+ "E",
+ "H"
+ ],
+ "direction": "vertical",
+ "origin": [
+ 200,
+ 215
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 0,
+ "fieldLabels": [
+ "Medium"
+ ]
+ },
+ "Roll": {
+ "fieldType": "QTYPE_INT",
+ "origin": [
+ 261,
+ 210
+ ],
+ "bubblesGap": 46.5,
+ "labelsGap": 58,
+ "fieldLabels": [
+ "roll1..9"
+ ]
+ },
+ "Int1": {
+ "fieldType": "QTYPE_INT",
+ "origin": [
+ 935,
+ 211
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 57,
+ "fieldLabels": [
+ "q6_1",
+ "q6_2"
+ ]
+ },
+ "Int2": {
+ "fieldType": "QTYPE_INT",
+ "origin": [
+ 1100,
+ 211
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 57,
+ "fieldLabels": [
+ "q7_1",
+ "q7_2"
+ ]
+ },
+ "Int3": {
+ "fieldType": "QTYPE_INT",
+ "origin": [
+ 1275,
+ 211
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 57,
+ "fieldLabels": [
+ "q8_1",
+ "q8_2"
+ ]
+ },
+ "Int4": {
+ "fieldType": "QTYPE_INT",
+ "origin": [
+ 1449,
+ 211
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 57,
+ "fieldLabels": [
+ "q9_1",
+ "q9_2"
+ ]
+ },
+ "Int5": {
+ "fieldType": "QTYPE_INT",
+ "origin": [
+ 1620,
+ 211
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 57,
+ "fieldLabels": [
+ "q10_1",
+ "q10_2"
+ ]
+ },
+ "Mcq1": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 198,
+ 826
+ ],
+ "bubblesGap": 93,
+ "labelsGap": 62,
+ "fieldLabels": [
+ "q1..5"
+ ]
+ },
+ "Mcq2": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 833,
+ 830
+ ],
+ "bubblesGap": 71,
+ "labelsGap": 61,
+ "fieldLabels": [
+ "q11..14"
+ ]
+ },
+ "Mcq3": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 833,
+ 1270
+ ],
+ "bubblesGap": 71,
+ "labelsGap": 61,
+ "fieldLabels": [
+ "q15..18"
+ ]
+ },
+ "Mcq4": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 1481,
+ 830
+ ],
+ "bubblesGap": 73,
+ "labelsGap": 61,
+ "fieldLabels": [
+ "q19..22"
+ ]
+ }
+ }
+}
diff --git a/samples/sample6/config.json b/samples/sample6/config.json
new file mode 100644
index 00000000..6f39f0ca
--- /dev/null
+++ b/samples/sample6/config.json
@@ -0,0 +1,11 @@
+{
+ "dimensions": {
+ "display_width": 2480,
+ "display_height": 3508,
+ "processing_width": 1653,
+ "processing_height": 2339
+ },
+ "outputs": {
+ "show_image_level": 5
+ }
+}
diff --git a/samples/sample6/doc-scans/sample_roll_01.jpg b/samples/sample6/doc-scans/sample_roll_01.jpg
new file mode 100644
index 00000000..25093a37
Binary files /dev/null and b/samples/sample6/doc-scans/sample_roll_01.jpg differ
diff --git a/samples/sample6/doc-scans/sample_roll_02.jpg b/samples/sample6/doc-scans/sample_roll_02.jpg
new file mode 100644
index 00000000..a81ee4fc
Binary files /dev/null and b/samples/sample6/doc-scans/sample_roll_02.jpg differ
diff --git a/samples/sample6/doc-scans/sample_roll_03.jpg b/samples/sample6/doc-scans/sample_roll_03.jpg
new file mode 100644
index 00000000..94296f4e
Binary files /dev/null and b/samples/sample6/doc-scans/sample_roll_03.jpg differ
diff --git a/samples/sample6/readme.md b/samples/sample6/readme.md
new file mode 100644
index 00000000..2a02ee90
--- /dev/null
+++ b/samples/sample6/readme.md
@@ -0,0 +1,21 @@
+# Demo for feature-based alignment
+
+## Background
+OMR is used to match student roll on final exam scripts. Scripts are scanned using a document scanner and the cover pages are extracted for OMR. Even though a document scanner does not produce any warpped perspective, the alignment is not perfect, causing some rotation and translation in the scans.
+
+The scripts in this sample were specifically selected incorrectly marked scripts to demonstrate how feature-based alignment can correct transformation errors using a reference image. In the actual batch. 156 out of 532 scripts were incorrectly marked. With feature-based alignment, all scripts were correctly marked.
+
+## Usage
+Two template files are given in the sample folder, one with feature-based alignment (template_fb_align), the other without (template_no_fb_align).
+
+## Additional Notes
+
+### Reference Image
+When using a reference image for feature-based alignment, it is better not to have many repeated patterns as it is causes ambiguity when trying to match similar feature points. The bubbles in an OMR form are identical and should not be used for feature-extraction.
+
+Thus, the reference image should be cleared of any bubbles. Forms with lots of text as in this example would be effective.
+
+Note the reference image in this sample was generated from a vector pdf, and not from a scanned blank, producing in a perfectly aligned reference.
+
+### Level adjustment
+The bubbles on the scripts were not shaded dark enough. Thus, a level adjustment was done to bring the black point to 70% to darken the light shading. White point was brought down to 80% to remove the light-grey background in the columns.
diff --git a/samples/sample6/reference.png b/samples/sample6/reference.png
new file mode 100644
index 00000000..5351267f
Binary files /dev/null and b/samples/sample6/reference.png differ
diff --git a/samples/sample6/template.json b/samples/sample6/template.json
new file mode 100644
index 00000000..57605a6c
--- /dev/null
+++ b/samples/sample6/template.json
@@ -0,0 +1,110 @@
+{
+ "pageDimensions": [
+ 2480,
+ 3508
+ ],
+ "bubbleDimensions": [
+ 42,
+ 42
+ ],
+ "preProcessors": [
+ {
+ "name": "Levels",
+ "options": {
+ "low": 0.7,
+ "high": 0.8
+ }
+ },
+ {
+ "name": "GaussianBlur",
+ "options": {
+ "kSize": [
+ 3,
+ 3
+ ],
+ "sigmaX": 0
+ }
+ }
+ ],
+ "customLabels": {
+ "Roll": [
+ "stu",
+ "roll1..7",
+ "check_1",
+ "check_2"
+ ]
+ },
+ "fieldBlocks": {
+ "Check1": {
+ "origin": [
+ 2033,
+ 1290
+ ],
+ "bubblesGap": 50,
+ "labelsGap": 50,
+ "fieldLabels": [
+ "check_1"
+ ],
+ "bubbleValues": [
+ "A",
+ "B",
+ "E",
+ "H",
+ "J",
+ "L",
+ "M"
+ ],
+ "direction": "vertical"
+ },
+ "Check2": {
+ "origin": [
+ 2083,
+ 1290
+ ],
+ "bubblesGap": 50,
+ "labelsGap": 50,
+ "fieldLabels": [
+ "check_2"
+ ],
+ "bubbleValues": [
+ "N",
+ "R",
+ "U",
+ "W",
+ "X",
+ "Y"
+ ],
+ "direction": "vertical"
+ },
+ "Stu": {
+ "origin": [
+ 1636,
+ 1290
+ ],
+ "bubblesGap": 50,
+ "labelsGap": 50,
+ "fieldLabels": [
+ "stu"
+ ],
+ "bubbleValues": [
+ "U",
+ "A",
+ "HT",
+ "GT"
+ ],
+ "direction": "vertical"
+ },
+ "Roll": {
+ "fieldType": "QTYPE_INT",
+ "origin": [
+ 1685,
+ 1290
+ ],
+ "bubblesGap": 50.5,
+ "labelsGap": 50.5,
+ "fieldLabels": [
+ "roll1..7"
+ ]
+ }
+ }
+}
diff --git a/samples/sample6/template_fb_align.json b/samples/sample6/template_fb_align.json
new file mode 100644
index 00000000..7c0c2397
--- /dev/null
+++ b/samples/sample6/template_fb_align.json
@@ -0,0 +1,118 @@
+{
+ "pageDimensions": [
+ 2480,
+ 3508
+ ],
+ "bubbleDimensions": [
+ 42,
+ 42
+ ],
+ "preProcessors": [
+ {
+ "name": "Levels",
+ "options": {
+ "low": 0.7,
+ "high": 0.8
+ }
+ },
+ {
+ "name": "FeatureBasedAlignment",
+ "options": {
+ "reference": "reference.png",
+ "maxFeatures": 1000,
+ "2d": true
+ }
+ },
+ {
+ "name": "GaussianBlur",
+ "options": {
+ "kSize": [
+ 3,
+ 3
+ ],
+ "sigmaX": 0
+ }
+ }
+ ],
+ "customLabels": {
+ "Roll": [
+ "stu",
+ "roll1..7",
+ "check_1",
+ "check_2"
+ ]
+ },
+ "fieldBlocks": {
+ "Check1": {
+ "origin": [
+ 2033,
+ 1290
+ ],
+ "bubblesGap": 50,
+ "labelsGap": 50,
+ "fieldLabels": [
+ "check_1"
+ ],
+ "bubbleValues": [
+ "A",
+ "B",
+ "E",
+ "H",
+ "J",
+ "L",
+ "M"
+ ],
+ "direction": "vertical"
+ },
+ "Check2": {
+ "origin": [
+ 2083,
+ 1290
+ ],
+ "bubblesGap": 50,
+ "labelsGap": 50,
+ "fieldLabels": [
+ "check_2"
+ ],
+ "bubbleValues": [
+ "N",
+ "R",
+ "U",
+ "W",
+ "X",
+ "Y"
+ ],
+ "direction": "vertical"
+ },
+ "Stu": {
+ "origin": [
+ 1636,
+ 1290
+ ],
+ "bubblesGap": 50,
+ "labelsGap": 50,
+ "fieldLabels": [
+ "stu"
+ ],
+ "bubbleValues": [
+ "U",
+ "A",
+ "HT",
+ "GT"
+ ],
+ "direction": "vertical"
+ },
+ "Roll": {
+ "fieldType": "QTYPE_INT",
+ "origin": [
+ 1685,
+ 1290
+ ],
+ "bubblesGap": 50.5,
+ "labelsGap": 50.5,
+ "fieldLabels": [
+ "roll1..7"
+ ]
+ }
+ }
+}
diff --git a/samples/sample6/template_no_fb_align.json b/samples/sample6/template_no_fb_align.json
new file mode 100644
index 00000000..57605a6c
--- /dev/null
+++ b/samples/sample6/template_no_fb_align.json
@@ -0,0 +1,110 @@
+{
+ "pageDimensions": [
+ 2480,
+ 3508
+ ],
+ "bubbleDimensions": [
+ 42,
+ 42
+ ],
+ "preProcessors": [
+ {
+ "name": "Levels",
+ "options": {
+ "low": 0.7,
+ "high": 0.8
+ }
+ },
+ {
+ "name": "GaussianBlur",
+ "options": {
+ "kSize": [
+ 3,
+ 3
+ ],
+ "sigmaX": 0
+ }
+ }
+ ],
+ "customLabels": {
+ "Roll": [
+ "stu",
+ "roll1..7",
+ "check_1",
+ "check_2"
+ ]
+ },
+ "fieldBlocks": {
+ "Check1": {
+ "origin": [
+ 2033,
+ 1290
+ ],
+ "bubblesGap": 50,
+ "labelsGap": 50,
+ "fieldLabels": [
+ "check_1"
+ ],
+ "bubbleValues": [
+ "A",
+ "B",
+ "E",
+ "H",
+ "J",
+ "L",
+ "M"
+ ],
+ "direction": "vertical"
+ },
+ "Check2": {
+ "origin": [
+ 2083,
+ 1290
+ ],
+ "bubblesGap": 50,
+ "labelsGap": 50,
+ "fieldLabels": [
+ "check_2"
+ ],
+ "bubbleValues": [
+ "N",
+ "R",
+ "U",
+ "W",
+ "X",
+ "Y"
+ ],
+ "direction": "vertical"
+ },
+ "Stu": {
+ "origin": [
+ 1636,
+ 1290
+ ],
+ "bubblesGap": 50,
+ "labelsGap": 50,
+ "fieldLabels": [
+ "stu"
+ ],
+ "bubbleValues": [
+ "U",
+ "A",
+ "HT",
+ "GT"
+ ],
+ "direction": "vertical"
+ },
+ "Roll": {
+ "fieldType": "QTYPE_INT",
+ "origin": [
+ 1685,
+ 1290
+ ],
+ "bubblesGap": 50.5,
+ "labelsGap": 50.5,
+ "fieldLabels": [
+ "roll1..7"
+ ]
+ }
+ }
+}
diff --git a/src/__init__.py b/src/__init__.py
new file mode 100644
index 00000000..7fc528b4
--- /dev/null
+++ b/src/__init__.py
@@ -0,0 +1,5 @@
+# https://docs.python.org/3/tutorial/modules.html#:~:text=The%20__init__.py,on%20the%20module%20search%20path.
+from src.logger import logger
+
+# It takes a few seconds for the imports
+logger.info(f"Loading OMRChecker modules...")
diff --git a/src/constants.py b/src/constants.py
new file mode 100644
index 00000000..f1cdc87b
--- /dev/null
+++ b/src/constants.py
@@ -0,0 +1,54 @@
+"""
+
+ OMRChecker
+
+ Author: Udayraj Deshmukh
+ Github: https://github.com/Udayraj123
+
+"""
+from dotmap import DotMap
+
+# Filenames
+TEMPLATE_FILENAME = "template.json"
+EVALUATION_FILENAME = "evaluation.json"
+CONFIG_FILENAME = "config.json"
+
+FIELD_LABEL_NUMBER_REGEX = r"([^\d]+)(\d*)"
+#
+ERROR_CODES = DotMap(
+ {
+ "MULTI_BUBBLE_WARN": 1,
+ "NO_MARKER_ERR": 2,
+ },
+ _dynamic=False,
+)
+
+FIELD_TYPES = {
+ "QTYPE_INT": {
+ "bubbleValues": ["0", "1", "2", "3", "4", "5", "6", "7", "8", "9"],
+ "direction": "vertical",
+ },
+ "QTYPE_INT_FROM_1": {
+ "bubbleValues": ["1", "2", "3", "4", "5", "6", "7", "8", "9", "0"],
+ "direction": "vertical",
+ },
+ "QTYPE_MCQ4": {"bubbleValues": ["A", "B", "C", "D"], "direction": "horizontal"},
+ "QTYPE_MCQ5": {
+ "bubbleValues": ["A", "B", "C", "D", "E"],
+ "direction": "horizontal",
+ },
+ #
+ # You can create and append custom field types here-
+ #
+}
+
+# TODO: move to interaction.py
+TEXT_SIZE = 0.95
+CLR_BLACK = (50, 150, 150)
+CLR_WHITE = (250, 250, 250)
+CLR_GRAY = (130, 130, 130)
+CLR_DARK_GRAY = (100, 100, 100)
+
+# TODO: move to config.json
+GLOBAL_PAGE_THRESHOLD_WHITE = 200
+GLOBAL_PAGE_THRESHOLD_BLACK = 100
diff --git a/src/core.py b/src/core.py
new file mode 100644
index 00000000..d475105f
--- /dev/null
+++ b/src/core.py
@@ -0,0 +1,721 @@
+import os
+from collections import defaultdict
+from typing import Any
+
+import cv2
+import matplotlib.pyplot as plt
+import numpy as np
+
+import src.constants as constants
+from src.logger import logger
+from src.utils.image import CLAHE_HELPER, ImageUtils
+from src.utils.interaction import InteractionUtils
+
+
+class ImageInstanceOps:
+ """Class to hold fine-tuned utilities for a group of images. One instance for each processing directory."""
+
+ save_img_list: Any = defaultdict(list)
+
+ def __init__(self, tuning_config):
+ super().__init__()
+ self.tuning_config = tuning_config
+ self.save_image_level = tuning_config.outputs.save_image_level
+
+ def apply_preprocessors(self, file_path, in_omr, template):
+ tuning_config = self.tuning_config
+ # resize to conform to template
+ in_omr = ImageUtils.resize_util(
+ in_omr,
+ tuning_config.dimensions.processing_width,
+ tuning_config.dimensions.processing_height,
+ )
+
+ # run pre_processors in sequence
+ for pre_processor in template.pre_processors:
+ in_omr = pre_processor.apply_filter(in_omr, file_path)
+ return in_omr
+
+ def read_omr_response(self, template, image, name, save_dir=None):
+ config = self.tuning_config
+ auto_align = config.alignment_params.auto_align
+ try:
+ img = image.copy()
+ # origDim = img.shape[:2]
+ img = ImageUtils.resize_util(
+ img, template.page_dimensions[0], template.page_dimensions[1]
+ )
+ if img.max() > img.min():
+ img = ImageUtils.normalize_util(img)
+ # Processing copies
+ transp_layer = img.copy()
+ final_marked = img.copy()
+
+ morph = img.copy()
+ self.append_save_img(3, morph)
+
+ if auto_align:
+ # Note: clahe is good for morphology, bad for thresholding
+ morph = CLAHE_HELPER.apply(morph)
+ self.append_save_img(3, morph)
+ # Remove shadows further, make columns/boxes darker (less gamma)
+ morph = ImageUtils.adjust_gamma(
+ morph, config.threshold_params.GAMMA_LOW
+ )
+ # TODO: all numbers should come from either constants or config
+ _, morph = cv2.threshold(morph, 220, 220, cv2.THRESH_TRUNC)
+ morph = ImageUtils.normalize_util(morph)
+ self.append_save_img(3, morph)
+ if config.outputs.show_image_level >= 4:
+ InteractionUtils.show("morph1", morph, 0, 1, config)
+
+ # Move them to data class if needed
+ # Overlay Transparencies
+ alpha = 0.65
+ omr_response = {}
+ multi_marked, multi_roll = 0, 0
+
+ # TODO Make this part useful for visualizing status checks
+ # blackVals=[0]
+ # whiteVals=[255]
+
+ if config.outputs.show_image_level >= 5:
+ all_c_box_vals = {"int": [], "mcq": []}
+ # TODO: simplify this logic
+ q_nums = {"int": [], "mcq": []}
+
+ # Find Shifts for the field_blocks --> Before calculating threshold!
+ if auto_align:
+ # print("Begin Alignment")
+ # Open : erode then dilate
+ v_kernel = cv2.getStructuringElement(cv2.MORPH_RECT, (2, 10))
+ morph_v = cv2.morphologyEx(
+ morph, cv2.MORPH_OPEN, v_kernel, iterations=3
+ )
+ _, morph_v = cv2.threshold(morph_v, 200, 200, cv2.THRESH_TRUNC)
+ morph_v = 255 - ImageUtils.normalize_util(morph_v)
+
+ if config.outputs.show_image_level >= 3:
+ InteractionUtils.show(
+ "morphed_vertical", morph_v, 0, 1, config=config
+ )
+
+ # InteractionUtils.show("morph1",morph,0,1,config=config)
+ # InteractionUtils.show("morphed_vertical",morph_v,0,1,config=config)
+
+ self.append_save_img(3, morph_v)
+
+ morph_thr = 60 # for Mobile images, 40 for scanned Images
+ _, morph_v = cv2.threshold(morph_v, morph_thr, 255, cv2.THRESH_BINARY)
+ # kernel best tuned to 5x5 now
+ morph_v = cv2.erode(morph_v, np.ones((5, 5), np.uint8), iterations=2)
+
+ self.append_save_img(3, morph_v)
+ # h_kernel = cv2.getStructuringElement(cv2.MORPH_RECT, (10, 2))
+ # morph_h = cv2.morphologyEx(morph, cv2.MORPH_OPEN, h_kernel, iterations=3)
+ # ret, morph_h = cv2.threshold(morph_h,200,200,cv2.THRESH_TRUNC)
+ # morph_h = 255 - normalize_util(morph_h)
+ # InteractionUtils.show("morph_h",morph_h,0,1,config=config)
+ # _, morph_h = cv2.threshold(morph_h,morph_thr,255,cv2.THRESH_BINARY)
+ # morph_h = cv2.erode(morph_h, np.ones((5,5),np.uint8), iterations = 2)
+ if config.outputs.show_image_level >= 3:
+ InteractionUtils.show(
+ "morph_thr_eroded", morph_v, 0, 1, config=config
+ )
+
+ self.append_save_img(6, morph_v)
+
+ # template relative alignment code
+ for field_block in template.field_blocks:
+ s, d = field_block.origin, field_block.dimensions
+
+ match_col, max_steps, align_stride, thk = map(
+ config.alignment_params.get,
+ [
+ "match_col",
+ "max_steps",
+ "stride",
+ "thickness",
+ ],
+ )
+ shift, steps = 0, 0
+ while steps < max_steps:
+ left_mean = np.mean(
+ morph_v[
+ s[1] : s[1] + d[1],
+ s[0] + shift - thk : -thk + s[0] + shift + match_col,
+ ]
+ )
+ right_mean = np.mean(
+ morph_v[
+ s[1] : s[1] + d[1],
+ s[0]
+ + shift
+ - match_col
+ + d[0]
+ + thk : thk
+ + s[0]
+ + shift
+ + d[0],
+ ]
+ )
+
+ # For demonstration purposes-
+ # if(field_block.name == "int1"):
+ # ret = morph_v.copy()
+ # cv2.rectangle(ret,
+ # (s[0]+shift-thk,s[1]),
+ # (s[0]+shift+thk+d[0],s[1]+d[1]),
+ # constants.CLR_WHITE,
+ # 3)
+ # appendSaveImg(6,ret)
+ # print(shift, left_mean, right_mean)
+ left_shift, right_shift = left_mean > 100, right_mean > 100
+ if left_shift:
+ if right_shift:
+ break
+ else:
+ shift -= align_stride
+ else:
+ if right_shift:
+ shift += align_stride
+ else:
+ break
+ steps += 1
+
+ field_block.shift = shift
+ # print("Aligned field_block: ",field_block.name,"Corrected Shift:",
+ # field_block.shift,", dimensions:", field_block.dimensions,
+ # "origin:", field_block.origin,'\n')
+ # print("End Alignment")
+
+ final_align = None
+ if config.outputs.show_image_level >= 2:
+ initial_align = self.draw_template_layout(img, template, shifted=False)
+ final_align = self.draw_template_layout(
+ img, template, shifted=True, draw_qvals=True
+ )
+ # appendSaveImg(4,mean_vals)
+ self.append_save_img(2, initial_align)
+ self.append_save_img(2, final_align)
+
+ if auto_align:
+ final_align = np.hstack((initial_align, final_align))
+ self.append_save_img(5, img)
+
+ # Get mean bubbleValues n other stats
+ all_q_vals, all_q_strip_arrs, all_q_std_vals = [], [], []
+ total_q_strip_no = 0
+ for field_block in template.field_blocks:
+ box_w, box_h = field_block.bubble_dimensions
+ q_std_vals = []
+ for field_block_bubbles in field_block.traverse_bubbles:
+ q_strip_vals = []
+ for pt in field_block_bubbles:
+ # shifted
+ x, y = (pt.x + field_block.shift, pt.y)
+ rect = [y, y + box_h, x, x + box_w]
+ q_strip_vals.append(
+ cv2.mean(img[rect[0] : rect[1], rect[2] : rect[3]])[0]
+ # detectCross(img, rect) ? 100 : 0
+ )
+ q_std_vals.append(round(np.std(q_strip_vals), 2))
+ all_q_strip_arrs.append(q_strip_vals)
+ # _, _, _ = get_global_threshold(q_strip_vals, "QStrip Plot",
+ # plot_show=False, sort_in_plot=True)
+ # hist = getPlotImg()
+ # InteractionUtils.show("QStrip "+field_block_bubbles[0].field_label, hist, 0, 1,config=config)
+ all_q_vals.extend(q_strip_vals)
+ # print(total_q_strip_no, field_block_bubbles[0].field_label, q_std_vals[len(q_std_vals)-1])
+ total_q_strip_no += 1
+ all_q_std_vals.extend(q_std_vals)
+
+ global_std_thresh, _, _ = self.get_global_threshold(
+ all_q_std_vals
+ ) # , "Q-wise Std-dev Plot", plot_show=True, sort_in_plot=True)
+ # plt.show()
+ # hist = getPlotImg()
+ # InteractionUtils.show("StdHist", hist, 0, 1,config=config)
+
+ # Note: Plotting takes Significant times here --> Change Plotting args
+ # to support show_image_level
+ # , "Mean Intensity Histogram",plot_show=True, sort_in_plot=True)
+ global_thr, _, _ = self.get_global_threshold(all_q_vals, looseness=4)
+
+ logger.info(
+ f"Thresholding: \tglobal_thr: {round(global_thr, 2)} \tglobal_std_THR: {round(global_std_thresh, 2)}\t{'(Looks like a Xeroxed OMR)' if (global_thr == 255) else ''}"
+ )
+ # plt.show()
+ # hist = getPlotImg()
+ # InteractionUtils.show("StdHist", hist, 0, 1,config=config)
+
+ # if(config.outputs.show_image_level>=1):
+ # hist = getPlotImg()
+ # InteractionUtils.show("Hist", hist, 0, 1,config=config)
+ # appendSaveImg(4,hist)
+ # appendSaveImg(5,hist)
+ # appendSaveImg(2,hist)
+
+ per_omr_threshold_avg, total_q_strip_no, total_q_box_no = 0, 0, 0
+ for field_block in template.field_blocks:
+ block_q_strip_no = 1
+ box_w, box_h = field_block.bubble_dimensions
+ shift = field_block.shift
+ s, d = field_block.origin, field_block.dimensions
+ key = field_block.name[:3]
+ # cv2.rectangle(final_marked,(s[0]+shift,s[1]),(s[0]+shift+d[0],
+ # s[1]+d[1]),CLR_BLACK,3)
+ for field_block_bubbles in field_block.traverse_bubbles:
+ # All Black or All White case
+ no_outliers = all_q_std_vals[total_q_strip_no] < global_std_thresh
+ # print(total_q_strip_no, field_block_bubbles[0].field_label,
+ # all_q_std_vals[total_q_strip_no], "no_outliers:", no_outliers)
+ per_q_strip_threshold = self.get_local_threshold(
+ all_q_strip_arrs[total_q_strip_no],
+ global_thr,
+ no_outliers,
+ f"Mean Intensity Histogram for {key}.{field_block_bubbles[0].field_label}.{block_q_strip_no}",
+ config.outputs.show_image_level >= 6,
+ )
+ # print(field_block_bubbles[0].field_label,key,block_q_strip_no, "THR: ",
+ # round(per_q_strip_threshold,2))
+ per_omr_threshold_avg += per_q_strip_threshold
+
+ # Note: Little debugging visualization - view the particular Qstrip
+ # if(
+ # 0
+ # # or "q17" in (field_block_bubbles[0].field_label)
+ # # or (field_block_bubbles[0].field_label+str(block_q_strip_no))=="q15"
+ # ):
+ # st, end = qStrip
+ # InteractionUtils.show("QStrip: "+key+"-"+str(block_q_strip_no),
+ # img[st[1] : end[1], st[0]+shift : end[0]+shift],0,config=config)
+
+ # TODO: get rid of total_q_box_no
+ detected_bubbles = []
+ for bubble in field_block_bubbles:
+ bubble_is_marked = (
+ per_q_strip_threshold > all_q_vals[total_q_box_no]
+ )
+ total_q_box_no += 1
+ if bubble_is_marked:
+ detected_bubbles.append(bubble)
+ x, y, field_value = (
+ bubble.x + field_block.shift,
+ bubble.y,
+ bubble.field_value,
+ )
+ cv2.rectangle(
+ final_marked,
+ (int(x + box_w / 12), int(y + box_h / 12)),
+ (
+ int(x + box_w - box_w / 12),
+ int(y + box_h - box_h / 12),
+ ),
+ constants.CLR_DARK_GRAY,
+ 3,
+ )
+
+ cv2.putText(
+ final_marked,
+ str(field_value),
+ (x, y),
+ cv2.FONT_HERSHEY_SIMPLEX,
+ constants.TEXT_SIZE,
+ (20, 20, 10),
+ int(1 + 3.5 * constants.TEXT_SIZE),
+ )
+ else:
+ cv2.rectangle(
+ final_marked,
+ (int(x + box_w / 10), int(y + box_h / 10)),
+ (
+ int(x + box_w - box_w / 10),
+ int(y + box_h - box_h / 10),
+ ),
+ constants.CLR_GRAY,
+ -1,
+ )
+
+ for bubble in detected_bubbles:
+ field_label, field_value = (
+ bubble.field_label,
+ bubble.field_value,
+ )
+ # Only send rolls multi-marked in the directory
+ multi_marked_local = field_label in omr_response
+ omr_response[field_label] = (
+ (omr_response[field_label] + field_value)
+ if multi_marked_local
+ else field_value
+ )
+ # TODO: generalize this into identifier
+ # multi_roll = multi_marked_local and "Roll" in str(q)
+ multi_marked = multi_marked or multi_marked_local
+
+ if len(detected_bubbles) == 0:
+ field_label = field_block_bubbles[0].field_label
+ omr_response[field_label] = field_block.empty_val
+
+ if config.outputs.show_image_level >= 5:
+ if key in all_c_box_vals:
+ q_nums[key].append(f"{key[:2]}_c{str(block_q_strip_no)}")
+ all_c_box_vals[key].append(
+ all_q_strip_arrs[total_q_strip_no]
+ )
+
+ block_q_strip_no += 1
+ total_q_strip_no += 1
+ # /for field_block
+
+ per_omr_threshold_avg /= total_q_strip_no
+ per_omr_threshold_avg = round(per_omr_threshold_avg, 2)
+ # Translucent
+ cv2.addWeighted(
+ final_marked, alpha, transp_layer, 1 - alpha, 0, final_marked
+ )
+ # Box types
+ if config.outputs.show_image_level >= 6:
+ # plt.draw()
+ f, axes = plt.subplots(len(all_c_box_vals), sharey=True)
+ f.canvas.manager.set_window_title(name)
+ ctr = 0
+ type_name = {
+ "int": "Integer",
+ "mcq": "MCQ",
+ "med": "MED",
+ "rol": "Roll",
+ }
+ for k, boxvals in all_c_box_vals.items():
+ axes[ctr].title.set_text(type_name[k] + " Type")
+ axes[ctr].boxplot(boxvals)
+ # thrline=axes[ctr].axhline(per_omr_threshold_avg,color='red',ls='--')
+ # thrline.set_label("Average THR")
+ axes[ctr].set_ylabel("Intensity")
+ axes[ctr].set_xticklabels(q_nums[k])
+ # axes[ctr].legend()
+ ctr += 1
+ # imshow will do the waiting
+ plt.tight_layout(pad=0.5)
+ plt.show()
+
+ if config.outputs.show_image_level >= 3 and final_align is not None:
+ final_align = ImageUtils.resize_util_h(
+ final_align, int(config.dimensions.display_height)
+ )
+ # [final_align.shape[1],0])
+ InteractionUtils.show(
+ "Template Alignment Adjustment", final_align, 0, 0, config=config
+ )
+
+ if config.outputs.save_detections and save_dir is not None:
+ if multi_roll:
+ save_dir = save_dir.joinpath("_MULTI_")
+ image_path = str(save_dir.joinpath(name))
+ ImageUtils.save_img(image_path, final_marked)
+
+ self.append_save_img(2, final_marked)
+
+ if save_dir is not None:
+ for i in range(config.outputs.save_image_level):
+ self.save_image_stacks(i + 1, name, save_dir)
+
+ return omr_response, final_marked, multi_marked, multi_roll
+
+ except Exception as e:
+ raise e
+
+ @staticmethod
+ def draw_template_layout(img, template, shifted=True, draw_qvals=False, border=-1):
+ img = ImageUtils.resize_util(
+ img, template.page_dimensions[0], template.page_dimensions[1]
+ )
+ final_align = img.copy()
+ for field_block in template.field_blocks:
+ s, d = field_block.origin, field_block.dimensions
+ box_w, box_h = field_block.bubble_dimensions
+ shift = field_block.shift
+ if shifted:
+ cv2.rectangle(
+ final_align,
+ (s[0] + shift, s[1]),
+ (s[0] + shift + d[0], s[1] + d[1]),
+ constants.CLR_BLACK,
+ 3,
+ )
+ else:
+ cv2.rectangle(
+ final_align,
+ (s[0], s[1]),
+ (s[0] + d[0], s[1] + d[1]),
+ constants.CLR_BLACK,
+ 3,
+ )
+ for field_block_bubbles in field_block.traverse_bubbles:
+ for pt in field_block_bubbles:
+ x, y = (pt.x + field_block.shift, pt.y) if shifted else (pt.x, pt.y)
+ cv2.rectangle(
+ final_align,
+ (int(x + box_w / 10), int(y + box_h / 10)),
+ (int(x + box_w - box_w / 10), int(y + box_h - box_h / 10)),
+ constants.CLR_GRAY,
+ border,
+ )
+ if draw_qvals:
+ rect = [y, y + box_h, x, x + box_w]
+ cv2.putText(
+ final_align,
+ f"{int(cv2.mean(img[rect[0] : rect[1], rect[2] : rect[3]])[0])}",
+ (rect[2] + 2, rect[0] + (box_h * 2) // 3),
+ cv2.FONT_HERSHEY_SIMPLEX,
+ 0.6,
+ constants.CLR_BLACK,
+ 2,
+ )
+ if shifted:
+ text_in_px = cv2.getTextSize(
+ field_block.name, cv2.FONT_HERSHEY_SIMPLEX, constants.TEXT_SIZE, 4
+ )
+ cv2.putText(
+ final_align,
+ field_block.name,
+ (int(s[0] + d[0] - text_in_px[0][0]), int(s[1] - text_in_px[0][1])),
+ cv2.FONT_HERSHEY_SIMPLEX,
+ constants.TEXT_SIZE,
+ constants.CLR_BLACK,
+ 4,
+ )
+ return final_align
+
+ def get_global_threshold(
+ self,
+ q_vals_orig,
+ plot_title=None,
+ plot_show=True,
+ sort_in_plot=True,
+ looseness=1,
+ ):
+ """
+ Note: Cannot assume qStrip has only-gray or only-white bg
+ (in which case there is only one jump).
+ So there will be either 1 or 2 jumps.
+ 1 Jump :
+ ......
+ ||||||
+ |||||| <-- risky THR
+ |||||| <-- safe THR
+ ....||||||
+ ||||||||||
+
+ 2 Jumps :
+ ......
+ |||||| <-- wrong THR
+ ....||||||
+ |||||||||| <-- safe THR
+ ..||||||||||
+ ||||||||||||
+
+ The abstract "First LARGE GAP" is perfect for this.
+ Current code is considering ONLY TOP 2 jumps(>= MIN_GAP) to be big,
+ gives the smaller one
+
+ """
+ config = self.tuning_config
+ PAGE_TYPE_FOR_THRESHOLD, MIN_JUMP, JUMP_DELTA = map(
+ config.threshold_params.get,
+ [
+ "PAGE_TYPE_FOR_THRESHOLD",
+ "MIN_JUMP",
+ "JUMP_DELTA",
+ ],
+ )
+
+ global_default_threshold = (
+ constants.GLOBAL_PAGE_THRESHOLD_WHITE
+ if PAGE_TYPE_FOR_THRESHOLD == "white"
+ else constants.GLOBAL_PAGE_THRESHOLD_BLACK
+ )
+
+ # Sort the Q bubbleValues
+ # TODO: Change var name of q_vals
+ q_vals = sorted(q_vals_orig)
+ # Find the FIRST LARGE GAP and set it as threshold:
+ ls = (looseness + 1) // 2
+ l = len(q_vals) - ls
+ max1, thr1 = MIN_JUMP, global_default_threshold
+ for i in range(ls, l):
+ jump = q_vals[i + ls] - q_vals[i - ls]
+ if jump > max1:
+ max1 = jump
+ thr1 = q_vals[i - ls] + jump / 2
+
+ # NOTE: thr2 is deprecated, thus is JUMP_DELTA
+ # Make use of the fact that the JUMP_DELTA(Vertical gap ofc) between
+ # values at detected jumps would be atleast 20
+ max2, thr2 = MIN_JUMP, global_default_threshold
+ # Requires atleast 1 gray box to be present (Roll field will ensure this)
+ for i in range(ls, l):
+ jump = q_vals[i + ls] - q_vals[i - ls]
+ new_thr = q_vals[i - ls] + jump / 2
+ if jump > max2 and abs(thr1 - new_thr) > JUMP_DELTA:
+ max2 = jump
+ thr2 = new_thr
+ # global_thr = min(thr1,thr2)
+ global_thr, j_low, j_high = thr1, thr1 - max1 // 2, thr1 + max1 // 2
+
+ # # For normal images
+ # thresholdRead = 116
+ # if(thr1 > thr2 and thr2 > thresholdRead):
+ # print("Note: taking safer thr line.")
+ # global_thr, j_low, j_high = thr2, thr2 - max2//2, thr2 + max2//2
+
+ if plot_title:
+ _, ax = plt.subplots()
+ ax.bar(range(len(q_vals_orig)), q_vals if sort_in_plot else q_vals_orig)
+ ax.set_title(plot_title)
+ thrline = ax.axhline(global_thr, color="green", ls="--", linewidth=5)
+ thrline.set_label("Global Threshold")
+ thrline = ax.axhline(thr2, color="red", ls=":", linewidth=3)
+ thrline.set_label("THR2 Line")
+ # thrline=ax.axhline(j_low,color='red',ls='-.', linewidth=3)
+ # thrline=ax.axhline(j_high,color='red',ls='-.', linewidth=3)
+ # thrline.set_label("Boundary Line")
+ # ax.set_ylabel("Mean Intensity")
+ ax.set_ylabel("Values")
+ ax.set_xlabel("Position")
+ ax.legend()
+ if plot_show:
+ plt.title(plot_title)
+ plt.show()
+
+ return global_thr, j_low, j_high
+
+ def get_local_threshold(
+ self, q_vals, global_thr, no_outliers, plot_title=None, plot_show=True
+ ):
+ """
+ TODO: Update this documentation too-
+ //No more - Assumption : Colwise background color is uniformly gray or white,
+ but not alternating. In this case there is atmost one jump.
+
+ 0 Jump :
+ <-- safe THR?
+ .......
+ ...|||||||
+ |||||||||| <-- safe THR?
+ // How to decide given range is above or below gray?
+ -> global q_vals shall absolutely help here. Just run same function
+ on total q_vals instead of colwise _//
+ How to decide it is this case of 0 jumps
+
+ 1 Jump :
+ ......
+ ||||||
+ |||||| <-- risky THR
+ |||||| <-- safe THR
+ ....||||||
+ ||||||||||
+
+ """
+ config = self.tuning_config
+ # Sort the Q bubbleValues
+ q_vals = sorted(q_vals)
+
+ # Small no of pts cases:
+ # base case: 1 or 2 pts
+ if len(q_vals) < 3:
+ thr1 = (
+ global_thr
+ if np.max(q_vals) - np.min(q_vals) < config.threshold_params.MIN_GAP
+ else np.mean(q_vals)
+ )
+ else:
+ # qmin, qmax, qmean, qstd = round(np.min(q_vals),2), round(np.max(q_vals),2),
+ # round(np.mean(q_vals),2), round(np.std(q_vals),2)
+ # GVals = [round(abs(q-qmean),2) for q in q_vals]
+ # gmean, gstd = round(np.mean(GVals),2), round(np.std(GVals),2)
+ # # DISCRETION: Pretty critical factor in reading response
+ # # Doesn't work well for small number of values.
+ # DISCRETION = 2.7 # 2.59 was closest hit, 3.0 is too far
+ # L2MaxGap = round(max([abs(g-gmean) for g in GVals]),2)
+ # if(L2MaxGap > DISCRETION*gstd):
+ # no_outliers = False
+
+ # # ^Stackoverflow method
+ # print(field_label, no_outliers,"qstd",round(np.std(q_vals),2), "gstd", gstd,
+ # "Gaps in gvals",sorted([round(abs(g-gmean),2) for g in GVals],reverse=True),
+ # '\t',round(DISCRETION*gstd,2), L2MaxGap)
+
+ # else:
+ # Find the LARGEST GAP and set it as threshold: //(FIRST LARGE GAP)
+ l = len(q_vals) - 1
+ max1, thr1 = config.threshold_params.MIN_JUMP, 255
+ for i in range(1, l):
+ jump = q_vals[i + 1] - q_vals[i - 1]
+ if jump > max1:
+ max1 = jump
+ thr1 = q_vals[i - 1] + jump / 2
+ # print(field_label,q_vals,max1)
+
+ confident_jump = (
+ config.threshold_params.MIN_JUMP
+ + config.threshold_params.CONFIDENT_SURPLUS
+ )
+ # If not confident, then only take help of global_thr
+ if max1 < confident_jump:
+ if no_outliers:
+ # All Black or All White case
+ thr1 = global_thr
+ else:
+ # TODO: Low confidence parameters here
+ pass
+
+ # if(thr1 == 255):
+ # print("Warning: threshold is unexpectedly 255! (Outlier Delta issue?)",plot_title)
+
+ # Make a common plot function to show local and global thresholds
+ if plot_show and plot_title is not None:
+ _, ax = plt.subplots()
+ ax.bar(range(len(q_vals)), q_vals)
+ thrline = ax.axhline(thr1, color="green", ls=("-."), linewidth=3)
+ thrline.set_label("Local Threshold")
+ thrline = ax.axhline(global_thr, color="red", ls=":", linewidth=5)
+ thrline.set_label("Global Threshold")
+ ax.set_title(plot_title)
+ ax.set_ylabel("Bubble Mean Intensity")
+ ax.set_xlabel("Bubble Number(sorted)")
+ ax.legend()
+ # TODO append QStrip to this plot-
+ # appendSaveImg(6,getPlotImg())
+ if plot_show:
+ plt.show()
+ return thr1
+
+ def append_save_img(self, key, img):
+ if self.save_image_level >= int(key):
+ self.save_img_list[key].append(img.copy())
+
+ def save_image_stacks(self, key, filename, save_dir):
+ config = self.tuning_config
+ if self.save_image_level >= int(key) and self.save_img_list[key] != []:
+ name = os.path.splitext(filename)[0]
+ result = np.hstack(
+ tuple(
+ [
+ ImageUtils.resize_util_h(img, config.dimensions.display_height)
+ for img in self.save_img_list[key]
+ ]
+ )
+ )
+ result = ImageUtils.resize_util(
+ result,
+ min(
+ len(self.save_img_list[key]) * config.dimensions.display_width // 3,
+ int(config.dimensions.display_width * 2.5),
+ ),
+ )
+ ImageUtils.save_img(f"{save_dir}stack/{name}_{str(key)}_stack.jpg", result)
+
+ def reset_all_save_img(self):
+ for i in range(self.save_image_level):
+ self.save_img_list[i + 1] = []
diff --git a/src/defaults/__init__.py b/src/defaults/__init__.py
new file mode 100644
index 00000000..62ed2e6f
--- /dev/null
+++ b/src/defaults/__init__.py
@@ -0,0 +1,5 @@
+# https://docs.python.org/3/tutorial/modules.html#:~:text=The%20__init__.py,on%20the%20module%20search%20path.
+# Use all imports relative to root directory
+# (https://chrisyeh96.github.io/2017/08/08/definitive-guide-python-imports.html)
+from src.defaults.config import * # NOQA
+from src.defaults.template import * # NOQA
diff --git a/src/defaults/config.py b/src/defaults/config.py
new file mode 100644
index 00000000..1c6cad3f
--- /dev/null
+++ b/src/defaults/config.py
@@ -0,0 +1,35 @@
+from dotmap import DotMap
+
+CONFIG_DEFAULTS = DotMap(
+ {
+ "dimensions": {
+ "display_height": 2480,
+ "display_width": 1640,
+ "processing_height": 820,
+ "processing_width": 666,
+ },
+ "threshold_params": {
+ "GAMMA_LOW": 0.7,
+ "MIN_GAP": 30,
+ "MIN_JUMP": 25,
+ "CONFIDENT_SURPLUS": 5,
+ "JUMP_DELTA": 30,
+ "PAGE_TYPE_FOR_THRESHOLD": "white",
+ },
+ "alignment_params": {
+ # Note: 'auto_align' enables automatic template alignment, use if the scans show slight misalignments.
+ "auto_align": False,
+ "match_col": 5,
+ "max_steps": 20,
+ "stride": 1,
+ "thickness": 3,
+ },
+ "outputs": {
+ "show_image_level": 0,
+ "save_image_level": 0,
+ "save_detections": True,
+ "filter_out_multimarked_files": False,
+ },
+ },
+ _dynamic=False,
+)
diff --git a/src/defaults/template.py b/src/defaults/template.py
new file mode 100644
index 00000000..d0a2a831
--- /dev/null
+++ b/src/defaults/template.py
@@ -0,0 +1,6 @@
+TEMPLATE_DEFAULTS = {
+ "preProcessors": [],
+ "emptyValue": "",
+ "customLabels": {},
+ "outputColumns": [],
+}
diff --git a/src/entry.py b/src/entry.py
new file mode 100644
index 00000000..5cfcfd95
--- /dev/null
+++ b/src/entry.py
@@ -0,0 +1,370 @@
+"""
+
+ OMRChecker
+
+ Author: Udayraj Deshmukh
+ Github: https://github.com/Udayraj123
+
+"""
+import os
+from csv import QUOTE_NONNUMERIC
+from pathlib import Path
+from time import time
+
+import cv2
+import pandas as pd
+from rich.table import Table
+
+from src import constants
+from src.defaults import CONFIG_DEFAULTS
+from src.evaluation import EvaluationConfig, evaluate_concatenated_response
+from src.logger import console, logger
+from src.template import Template
+from src.utils.file import Paths, setup_dirs_for_paths, setup_outputs_for_template
+from src.utils.image import ImageUtils
+from src.utils.interaction import InteractionUtils, Stats
+from src.utils.parsing import get_concatenated_response, open_config_with_defaults
+
+# Load processors
+STATS = Stats()
+
+
+def entry_point(input_dir, args):
+ if not os.path.exists(input_dir):
+ raise Exception(f"Given input directory does not exist: '{input_dir}'")
+ curr_dir = input_dir
+ return process_dir(input_dir, curr_dir, args)
+
+
+def print_config_summary(
+ curr_dir,
+ omr_files,
+ template,
+ tuning_config,
+ local_config_path,
+ evaluation_config,
+ args,
+):
+ logger.info("")
+ table = Table(title="Current Configurations", show_header=False, show_lines=False)
+ table.add_column("Key", style="cyan", no_wrap=True)
+ table.add_column("Value", style="magenta")
+ table.add_row("Directory Path", f"{curr_dir}")
+ table.add_row("Count of Images", f"{len(omr_files)}")
+ table.add_row("Set Layout Mode ", "ON" if args["setLayout"] else "OFF")
+ table.add_row(
+ "Markers Detection",
+ "ON" if "CropOnMarkers" in template.pre_processors else "OFF",
+ )
+ table.add_row("Auto Alignment", f"{tuning_config.alignment_params.auto_align}")
+ table.add_row("Detected Template Path", f"{template}")
+ if local_config_path:
+ table.add_row("Detected Local Config", f"{local_config_path}")
+ if evaluation_config:
+ table.add_row("Detected Evaluation Config", f"{evaluation_config}")
+
+ table.add_row(
+ "Detected pre-processors",
+ f"{[pp.__class__.__name__ for pp in template.pre_processors]}",
+ )
+ console.print(table, justify="center")
+
+
+def process_dir(
+ root_dir,
+ curr_dir,
+ args,
+ template=None,
+ tuning_config=CONFIG_DEFAULTS,
+ evaluation_config=None,
+):
+ # Update local tuning_config (in current recursion stack)
+ local_config_path = curr_dir.joinpath(constants.CONFIG_FILENAME)
+ if os.path.exists(local_config_path):
+ tuning_config = open_config_with_defaults(local_config_path)
+
+ # Update local template (in current recursion stack)
+ local_template_path = curr_dir.joinpath(constants.TEMPLATE_FILENAME)
+ local_template_exists = os.path.exists(local_template_path)
+ if local_template_exists:
+ template = Template(
+ local_template_path,
+ tuning_config,
+ )
+ # Look for subdirectories for processing
+ subdirs = [d for d in curr_dir.iterdir() if d.is_dir()]
+
+ output_dir = Path(args["output_dir"], curr_dir.relative_to(root_dir))
+ paths = Paths(output_dir)
+
+ # look for images in current dir to process
+ exts = ("*.[pP][nN][gG]", "*.[jJ][pP][gG]", "*.[jJ][pP][eE][gG]")
+ omr_files = sorted([f for ext in exts for f in curr_dir.glob(ext)])
+
+ # Exclude images (take union over all pre_processors)
+ excluded_files = []
+ if template:
+ for pp in template.pre_processors:
+ excluded_files.extend(Path(p) for p in pp.exclude_files())
+
+ local_evaluation_path = curr_dir.joinpath(constants.EVALUATION_FILENAME)
+ if not args["setLayout"] and os.path.exists(local_evaluation_path):
+ if not local_template_exists:
+ logger.warning(
+ f"Found an evaluation file without a parent template file: {local_evaluation_path}"
+ )
+ evaluation_config = EvaluationConfig(
+ curr_dir,
+ local_evaluation_path,
+ template,
+ tuning_config,
+ )
+
+ excluded_files.extend(
+ Path(exclude_file) for exclude_file in evaluation_config.get_exclude_files()
+ )
+
+ omr_files = [f for f in omr_files if f not in excluded_files]
+
+ if omr_files:
+ if not template:
+ logger.error(
+ f"Found images, but no template in the directory tree \
+ of '{curr_dir}'. \nPlace {constants.TEMPLATE_FILENAME} in the \
+ appropriate directory."
+ )
+ raise Exception(
+ f"No template file found in the directory tree of {curr_dir}"
+ )
+
+ setup_dirs_for_paths(paths)
+ outputs_namespace = setup_outputs_for_template(paths, template)
+
+ print_config_summary(
+ curr_dir,
+ omr_files,
+ template,
+ tuning_config,
+ local_config_path,
+ evaluation_config,
+ args,
+ )
+ if args["setLayout"]:
+ show_template_layouts(omr_files, template, tuning_config)
+ else:
+ process_files(
+ omr_files,
+ template,
+ tuning_config,
+ evaluation_config,
+ outputs_namespace,
+ )
+
+ elif not subdirs:
+ # Each subdirectory should have images or should be non-leaf
+ logger.info(
+ f"No valid images or sub-folders found in {curr_dir}.\
+ Empty directories not allowed."
+ )
+
+ # recursively process sub-folders
+ for d in subdirs:
+ process_dir(
+ root_dir,
+ d,
+ args,
+ template,
+ tuning_config,
+ evaluation_config,
+ )
+
+
+def show_template_layouts(omr_files, template, tuning_config):
+ for file_path in omr_files:
+ file_name = file_path.name
+ file_path = str(file_path)
+ in_omr = cv2.imread(file_path, cv2.IMREAD_GRAYSCALE)
+ in_omr = template.image_instance_ops.apply_preprocessors(
+ file_path, in_omr, template
+ )
+ template_layout = template.image_instance_ops.draw_template_layout(
+ in_omr, template, shifted=False, border=2
+ )
+ InteractionUtils.show(
+ f"Template Layout: {file_name}", template_layout, 1, 1, config=tuning_config
+ )
+
+
+def process_files(
+ omr_files,
+ template,
+ tuning_config,
+ evaluation_config,
+ outputs_namespace,
+):
+ start_time = int(time())
+ files_counter = 0
+ STATS.files_not_moved = 0
+
+ for file_path in omr_files:
+ files_counter += 1
+ file_name = file_path.name
+
+ in_omr = cv2.imread(str(file_path), cv2.IMREAD_GRAYSCALE)
+
+ logger.info("")
+ logger.info(
+ f"({files_counter}) Opening image: \t'{file_path}'\tResolution: {in_omr.shape}"
+ )
+
+ template.image_instance_ops.reset_all_save_img()
+
+ template.image_instance_ops.append_save_img(1, in_omr)
+
+ in_omr = template.image_instance_ops.apply_preprocessors(
+ file_path, in_omr, template
+ )
+
+ if in_omr is None:
+ # Error OMR case
+ new_file_path = outputs_namespace.paths.errors_dir.joinpath(file_name)
+ outputs_namespace.OUTPUT_SET.append(
+ [file_name] + outputs_namespace.empty_resp
+ )
+ if check_and_move(
+ constants.ERROR_CODES.NO_MARKER_ERR, file_path, new_file_path
+ ):
+ err_line = [
+ file_name,
+ file_path,
+ new_file_path,
+ "NA",
+ ] + outputs_namespace.empty_resp
+ pd.DataFrame(err_line, dtype=str).T.to_csv(
+ outputs_namespace.files_obj["Errors"],
+ mode="a",
+ quoting=QUOTE_NONNUMERIC,
+ header=False,
+ index=False,
+ )
+ continue
+
+ # uniquify
+ file_id = str(file_name)
+ save_dir = outputs_namespace.paths.save_marked_dir
+ (
+ response_dict,
+ final_marked,
+ multi_marked,
+ _,
+ ) = template.image_instance_ops.read_omr_response(
+ template, image=in_omr, name=file_id, save_dir=save_dir
+ )
+
+ # TODO: move inner try catch here
+ # concatenate roll nos, set unmarked responses, etc
+ omr_response = get_concatenated_response(response_dict, template)
+
+ if (
+ evaluation_config is None
+ or not evaluation_config.get_should_explain_scoring()
+ ):
+ logger.info(f"Read Response: \n{omr_response}")
+
+ score = 0
+ if evaluation_config is not None:
+ score = evaluate_concatenated_response(
+ omr_response, evaluation_config, file_path, outputs_namespace.paths.evaluation_dir
+ )
+ logger.info(
+ f"(/{files_counter}) Graded with score: {round(score, 2)}\t for file: '{file_id}'"
+ )
+ else:
+ logger.info(f"(/{files_counter}) Processed file: '{file_id}'")
+
+ if tuning_config.outputs.show_image_level >= 2:
+ InteractionUtils.show(
+ f"Final Marked Bubbles : '{file_id}'",
+ ImageUtils.resize_util_h(
+ final_marked, int(tuning_config.dimensions.display_height * 1.3)
+ ),
+ 1,
+ 1,
+ config=tuning_config,
+ )
+
+ resp_array = []
+ for k in template.output_columns:
+ resp_array.append(omr_response[k])
+
+ outputs_namespace.OUTPUT_SET.append([file_name] + resp_array)
+
+ if multi_marked == 0 or not tuning_config.outputs.filter_out_multimarked_files:
+ STATS.files_not_moved += 1
+ new_file_path = save_dir.joinpath(file_id)
+ # Enter into Results sheet-
+ results_line = [file_name, file_path, new_file_path, score] + resp_array
+ # Write/Append to results_line file(opened in append mode)
+ pd.DataFrame(results_line, dtype=str).T.to_csv(
+ outputs_namespace.files_obj["Results"],
+ mode="a",
+ quoting=QUOTE_NONNUMERIC,
+ header=False,
+ index=False,
+ )
+ else:
+ # multi_marked file
+ logger.info(f"[{files_counter}] Found multi-marked file: '{file_id}'")
+ new_file_path = outputs_namespace.paths.multi_marked_dir.joinpath(file_name)
+ if check_and_move(
+ constants.ERROR_CODES.MULTI_BUBBLE_WARN, file_path, new_file_path
+ ):
+ mm_line = [file_name, file_path, new_file_path, "NA"] + resp_array
+ pd.DataFrame(mm_line, dtype=str).T.to_csv(
+ outputs_namespace.files_obj["MultiMarked"],
+ mode="a",
+ quoting=QUOTE_NONNUMERIC,
+ header=False,
+ index=False,
+ )
+ # else:
+ # TODO: Add appropriate record handling here
+ # pass
+
+ print_stats(start_time, files_counter, tuning_config)
+
+
+def check_and_move(error_code, file_path, filepath2):
+ # TODO: fix file movement into error/multimarked/invalid etc again
+ STATS.files_not_moved += 1
+ return True
+
+
+def print_stats(start_time, files_counter, tuning_config):
+ time_checking = max(1, round(time() - start_time, 2))
+ log = logger.info
+ log("")
+ log(f"{'Total file(s) moved': <27}: {STATS.files_moved}")
+ log(f"{'Total file(s) not moved': <27}: {STATS.files_not_moved}")
+ log("--------------------------------")
+ log(
+ f"{'Total file(s) processed': <27}: {files_counter} ({'Sum Tallied!' if files_counter == (STATS.files_moved + STATS.files_not_moved) else 'Not Tallying!'})"
+ )
+
+ if tuning_config.outputs.show_image_level <= 0:
+ log(
+ f"\nFinished Checking {files_counter} file(s) in {round(time_checking, 1)} seconds i.e. ~{round(time_checking / 60, 1)} minute(s)."
+ )
+ log(
+ f"{'OMR Processing Rate': <27}: \t ~ {round(time_checking / files_counter, 2)} seconds/OMR"
+ )
+ log(
+ f"{'OMR Processing Speed': <27}: \t ~ {round((files_counter * 60) / time_checking, 2)} OMRs/minute"
+ )
+ else:
+ log(f"\n{'Total script time': <27}: {time_checking} seconds")
+
+ if tuning_config.outputs.show_image_level <= 1:
+ log(
+ "\nTip: To see some awesome visuals, open config.json and increase 'show_image_level'"
+ )
diff --git a/src/evaluation.py b/src/evaluation.py
new file mode 100644
index 00000000..0b567169
--- /dev/null
+++ b/src/evaluation.py
@@ -0,0 +1,546 @@
+import ast
+import os
+import re
+from copy import deepcopy
+from csv import QUOTE_NONNUMERIC
+
+import cv2
+import pandas as pd
+from rich.table import Table
+
+from src.logger import console, logger
+from src.schemas.constants import (
+ BONUS_SECTION_PREFIX,
+ DEFAULT_SECTION_KEY,
+ MARKING_VERDICT_TYPES,
+)
+from src.utils.parsing import (
+ get_concatenated_response,
+ open_evaluation_with_validation,
+ parse_fields,
+ parse_float_or_fraction,
+)
+
+
+class AnswerMatcher:
+ def __init__(self, answer_item, section_marking_scheme):
+ self.section_marking_scheme = section_marking_scheme
+ self.answer_item = answer_item
+ self.answer_type = self.validate_and_get_answer_type(answer_item)
+ self.set_defaults_from_scheme(section_marking_scheme)
+
+ @staticmethod
+ def is_a_marking_score(answer_element):
+ # Note: strict type checking is already done at schema validation level,
+ # Here we focus on overall struct type
+ return type(answer_element) == str or type(answer_element) == int
+
+ @staticmethod
+ def is_standard_answer(answer_element):
+ return type(answer_element) == str and len(answer_element) >= 1
+
+ def validate_and_get_answer_type(self, answer_item):
+ if self.is_standard_answer(answer_item):
+ return "standard"
+ elif type(answer_item) == list:
+ if (
+ # Array of answer elements: ['A', 'B', 'AB']
+ len(answer_item) >= 2
+ and all(
+ self.is_standard_answer(answers_or_score)
+ for answers_or_score in answer_item
+ )
+ ):
+ return "multiple-correct"
+ elif (
+ # Array of two-tuples: [['A', 1], ['B', 1], ['C', 3], ['AB', 2]]
+ len(answer_item) >= 1
+ and all(
+ type(answer_and_score) == list and len(answer_and_score) == 2
+ for answer_and_score in answer_item
+ )
+ and all(
+ self.is_standard_answer(allowed_answer)
+ and self.is_a_marking_score(answer_score)
+ for allowed_answer, answer_score in answer_item
+ )
+ ):
+ return "multiple-correct-weighted"
+
+ logger.critical(
+ f"Unable to determine answer type for answer item: {answer_item}"
+ )
+ raise Exception("Unable to determine answer type")
+
+ def set_defaults_from_scheme(self, section_marking_scheme):
+ answer_type = self.answer_type
+ self.empty_val = section_marking_scheme.empty_val
+ answer_item = self.answer_item
+ self.marking = deepcopy(section_marking_scheme.marking)
+ # TODO: reuse part of parse_scheme_marking here -
+ if answer_type == "standard":
+ # no local overrides
+ pass
+ elif answer_type == "multiple-correct":
+ # override marking scheme scores for each allowed answer
+ for allowed_answer in answer_item:
+ self.marking[f"correct-{allowed_answer}"] = self.marking["correct"]
+ elif answer_type == "multiple-correct-weighted":
+ # Note: No override using marking scheme as answer scores are provided in answer_item
+ for allowed_answer, answer_score in answer_item:
+ self.marking[f"correct-{allowed_answer}"] = parse_float_or_fraction(
+ answer_score
+ )
+
+ def get_marking_scheme(self):
+ return self.section_marking_scheme
+
+ def get_section_explanation(self):
+ answer_type = self.answer_type
+ if answer_type in ["standard", "multiple-correct"]:
+ return self.section_marking_scheme.section_key
+ elif answer_type == "multiple-correct-weighted":
+ return f"Custom: {self.marking}"
+
+ def get_verdict_marking(self, marked_answer):
+ answer_type = self.answer_type
+ question_verdict = "incorrect"
+ if answer_type == "standard":
+ question_verdict = self.get_standard_verdict(marked_answer)
+ elif answer_type == "multiple-correct":
+ question_verdict = self.get_multiple_correct_verdict(marked_answer)
+ elif answer_type == "multiple-correct-weighted":
+ question_verdict = self.get_multiple_correct_weighted_verdict(marked_answer)
+ return question_verdict, self.marking[question_verdict]
+
+ def get_standard_verdict(self, marked_answer):
+ allowed_answer = self.answer_item
+ if marked_answer == self.empty_val:
+ return "unmarked"
+ elif marked_answer == allowed_answer:
+ return "correct"
+ else:
+ return "incorrect"
+
+ def get_multiple_correct_verdict(self, marked_answer):
+ allowed_answers = self.answer_item
+ if marked_answer == self.empty_val:
+ return "unmarked"
+ elif marked_answer in allowed_answers:
+ return f"correct-{marked_answer}"
+ else:
+ return "incorrect"
+
+ def get_multiple_correct_weighted_verdict(self, marked_answer):
+ allowed_answers = [
+ allowed_answer for allowed_answer, _answer_score in self.answer_item
+ ]
+ if marked_answer == self.empty_val:
+ return "unmarked"
+ elif marked_answer in allowed_answers:
+ return f"correct-{marked_answer}"
+ else:
+ return "incorrect"
+
+ def __str__(self):
+ return f"{self.answer_item}"
+
+
+class SectionMarkingScheme:
+ def __init__(self, section_key, section_scheme, empty_val):
+ # TODO: get local empty_val from qblock
+ self.empty_val = empty_val
+ self.section_key = section_key
+ # DEFAULT marking scheme follows a shorthand
+ if section_key == DEFAULT_SECTION_KEY:
+ self.questions = None
+ self.marking = self.parse_scheme_marking(section_scheme)
+ else:
+ self.questions = parse_fields(section_key, section_scheme["questions"])
+ self.marking = self.parse_scheme_marking(section_scheme["marking"])
+
+ def __str__(self):
+ return self.section_key
+
+ def parse_scheme_marking(self, marking):
+ parsed_marking = {}
+ for verdict_type in MARKING_VERDICT_TYPES:
+ verdict_marking = parse_float_or_fraction(marking[verdict_type])
+ if (
+ verdict_marking > 0
+ and verdict_type == "incorrect"
+ and not self.section_key.startswith(BONUS_SECTION_PREFIX)
+ ):
+ logger.warning(
+ f"Found positive marks({round(verdict_marking, 2)}) for incorrect answer in the schema '{self.section_key}'. For Bonus sections, add a prefix 'BONUS_' to them."
+ )
+ parsed_marking[verdict_type] = verdict_marking
+
+ return parsed_marking
+
+ def match_answer(self, marked_answer, answer_matcher):
+ question_verdict, verdict_marking = answer_matcher.get_verdict_marking(
+ marked_answer
+ )
+
+ return verdict_marking, question_verdict
+
+
+class EvaluationConfig:
+ """Note: this instance will be reused for multiple omr sheets"""
+
+ def __init__(self, curr_dir, evaluation_path, template, tuning_config):
+ self.path = evaluation_path
+ evaluation_json = open_evaluation_with_validation(evaluation_path)
+ options, marking_schemes, source_type = map(
+ evaluation_json.get, ["options", "marking_schemes", "source_type"]
+ )
+ self.should_explain_scoring = options.get("should_explain_scoring", False)
+ self.has_non_default_section = False
+ self.exclude_files = []
+ self.enable_evaluation_table_to_csv = options.get(
+ "enable_evaluation_table_to_csv", False
+ )
+
+ if source_type == "csv":
+ csv_path = curr_dir.joinpath(options["answer_key_csv_path"])
+ if not os.path.exists(csv_path):
+ logger.warning(f"Answer key csv does not exist at: '{csv_path}'.")
+
+ answer_key_image_path = options.get("answer_key_image_path", None)
+ if os.path.exists(csv_path):
+ # TODO: CSV parsing/validation for each row with a (qNo,
[](https://www.paypal.me/Udayraj123/500)
+
+_Find OMRChecker on_ [**_Product Hunt_**](https://www.producthunt.com/posts/omr-checker/) **|** [**_Reddit_**](https://www.reddit.com/r/computervision/comments/ccbj6f/omrchecker_grade_exams_using_python_and_opencv/) **|** [**Discord**](https://discord.gg/qFv2Vqf) **|** [**Linkedin**](https://www.linkedin.com/pulse/open-source-talks-udayraj-udayraj-deshmukh/) **|** [**goodfirstissue.dev**](https://goodfirstissue.dev/language/python) **|** [**codepeak.tech**](https://www.codepeak.tech/) **|** [**fossoverflow.dev**](https://fossoverflow.dev/projects) **|** [**Interview on Console by CodeSee**](https://console.substack.com/p/console-140) **|** [**Open Source Hub**](https://opensourcehub.io/udayraj123/omrchecker)
-
-*Find OMRChecker on* [***Product Hunt***](https://www.producthunt.com/posts/omr-checker/) **|** [***Hacker News***](https://news.ycombinator.com/item?id=20420602) **|** [***Reddit***](https://www.reddit.com/r/computervision/comments/ccbj6f/omrchecker_grade_exams_using_python_and_opencv/) **|** [***Swyya***](https://www.swyya.com/projects/omrchecker) **|** [](https://discord.gg/qFv2Vqf)
+
+
diff --git a/docs/assets/colored_output.jpg b/docs/assets/colored_output.jpg
new file mode 100644
index 00000000..3cafa473
Binary files /dev/null and b/docs/assets/colored_output.jpg differ
diff --git a/globals.py b/globals.py
deleted file mode 100644
index 5f9cd929..00000000
--- a/globals.py
+++ /dev/null
@@ -1,140 +0,0 @@
-"""
-
-Designed and Developed by-
-Udayraj Deshmukh
-https://github.com/Udayraj123
-
-"""
-
-"""
-Constants
-"""
-display_height = int(480)
-display_width = int(640)
-windowWidth = 1280
-windowHeight = 720
-
-saveMarked = 1
-saveCropped = 1
-showimglvl = 4
-saveimglvl = 0
-PRELIM_CHECKS = 0
-saveImgList = {}
-resetpos = [0, 0]
-explain = 0
-# autorotate=1
-
-BATCH_NO = 1000
-NO_MARKER_ERR = 12
-MULTI_BUBBLE_WARN = 15
-
-# name of template file
-TEMPLATE_FILE = 'template.json'
-MARKER_FILE = "omr_marker.jpg"
-
-# For preProcessing
-GAMMA_LOW = 0.7
-GAMMA_HIGH = 1.25
-
-ERODE_SUB_OFF = 1
-
-# For new ways of determining threshold
-MIN_GAP, MIN_STD = 30, 25
-MIN_JUMP = 25
-# If only not confident, take help of globalTHR
-CONFIDENT_JUMP = MIN_JUMP + 15
-JUMP_DELTA = 30
-# MIN_GAP : worst case gap of black and gray
-
-# Templ alignment parameters
-ALIGN_RANGE = range(-5, 6, 1)
-# TODO ^THIS SHOULD BE IN LAYOUT FILE AS ITS RELATED TO DIMENSIONS
-# ALIGN_RANGE = [-6,-4,-2,-1,0,1,2,4,6]
-
-# max threshold difference for template matching
-thresholdVar = 0.41
-
-# TODO: remove unnec variables here-
-thresholdCircle = 0.3
-marker_rescale_range = (35, 100)
-marker_rescale_steps = 10
-
-# Presentation variables
-uniform_height = int(1231 / 1.5)
-uniform_width = int(1000 / 1.5)
-# Original dims are about (3527, 2494)
-
-# Any input images should be resized to this--
-uniform_width_hd = int(uniform_width * 1.5)
-uniform_height_hd = int(uniform_height * 1.5)
-
-TEXT_SIZE = 0.95
-CLR_BLACK = (50, 150, 150)
-CLR_WHITE = (250, 250, 250)
-CLR_GRAY = (130, 130, 130)
-# CLR_DARK_GRAY = (190,190,190)
-CLR_DARK_GRAY = (100, 100, 100)
-
-MIN_PAGE_AREA = 80000
-
-# Filepaths
-
-
-class Paths:
- def __init__(self, output):
- self.output = output
- self.saveMarkedDir = f'{output}/CheckedOMRs/'
- self.resultDir = f'{output}/Results/'
- self.manualDir = f'{output}/Manual/'
- self.errorsDir = f'{self.manualDir}ErrorFiles/'
- self.badRollsDir = f'{self.manualDir}BadRollNosFiles/'
- self.multiMarkedDir = f'{self.manualDir}MultiMarkedFiles/'
-
-
-"""
-Variables
-"""
-filesMoved = 0
-filesNotMoved = 0
-
-# for positioning image windows
-windowX, windowY = 0, 0
-
-
-# TODO: move to template or similar json
-Answers = {
- 'J': {
- 'q1': ['B'], 'q2': ['B'], 'q3': ['B'], 'q4': ['C'], 'q5': ['0', '00'], 'q6': ['0', '00'], 'q7': ['4', '04'],
- 'q8': ['9', '09'], 'q9': ['11', '11'], 'q10': ['C'], 'q11': ['C'], 'q12': ['B'], 'q13': ['C'],
- 'q14': ['C'], 'q15': ['B'], 'q16': ['C'], 'q17': ['BONUS'], 'q18': ['A'], 'q19': ['C'], 'q20': ['B']},
- 'H': {
- 'q1': ['B'], 'q2': ['BONUS'], 'q3': ['A'], 'q4': ['B'], 'q5': ['A'], 'q6': ['B'], 'q7': ['B'],
- 'q8': ['C'], 'q9': ['4', '04'], 'q10': ['4', '04'], 'q11': ['5', '05'], 'q12': ['1', '01'], 'q13': ['28'],
- 'q14': ['C'], 'q15': ['B'], 'q16': ['C'], 'q17': ['C'], 'q18': ['C'], 'q19': ['B'], 'q20': ['C']},
- 'JK': {
- 'q1': ['B'], 'q2': ['B'], 'q3': ['B'], 'q4': ['C'], 'q5': ['0', '00'], 'q6': ['0', '00'], 'q7': ['4', '04'],
- 'q8': ['9', '09'], 'q9': ['11', '11'], 'q10': ['C'], 'q11': ['C'], 'q12': ['B'], 'q13': ['C'],
- 'q14': ['C'], 'q15': ['B'], 'q16': ['C'], 'q17': ['BONUS'], 'q18': ['A'], 'q19': ['C'], 'q20': ['B']},
- 'HK': {
- 'q1': ['B'], 'q2': ['BONUS'], 'q3': ['A'], 'q4': ['B'], 'q5': ['B'], 'q6': ['B'], 'q7': ['B'],
- 'q8': ['C'], 'q9': ['4', '04'], 'q10': ['4', '04'], 'q11': ['5', '05'], 'q12': ['1', '01'], 'q13': ['28'],
- 'q14': ['C'], 'q15': ['B'], 'q16': ['C'], 'q17': ['C'], 'q18': ['C'], 'q19': ['B'], 'q20': ['C']},
-}
-
-# TODO: Make this generalized and move it to samples
-Sections = {
- 'J': {
- 'Fibo1': {'ques': [1, 2, 3, 4], '+seq': [2, 3, 5, 8], '-seq': [0, 1, 1, 2]},
- 'Power1': {'ques': [5, 6, 7, 8, 9], '+seq': [1, 2, 4, 8, 16], '-seq': [0, 0, 0, 0, 0]},
- 'Fibo2': {'ques': [10, 11, 12, 13], '+seq': [2, 3, 5, 8], '-seq': [0, 1, 1, 2]},
- 'allNone1': {'ques': [14, 15, 16], 'marks': 12},
- 'Boom1': {'ques': [17, 18, 19, 20], '+seq': [3, 3, 3, 3], '-seq': [1, 1, 1, 1]},
- },
- 'H': {
- 'Boom1': {'ques': [1, 2, 3, 4], '+seq': [3, 3, 3, 3], '-seq': [1, 1, 1, 1]},
- 'Fibo1': {'ques': [5, 6, 7, 8], '+seq': [2, 3, 5, 8], '-seq': [0, 1, 1, 2]},
- 'Power1': {'ques': [9, 10, 11, 12, 13], '+seq': [1, 2, 4, 8, 16], '-seq': [0, 0, 0, 0, 0]},
- 'allNone1': {'ques': [14, 15, 16], 'marks': 12},
- 'Boom2': {'ques': [17, 18, 19, 20], '+seq': [3, 3, 3, 3], '-seq': [1, 1, 1, 1]},
- },
-}
diff --git a/main.py b/main.py
index eac70a50..dfecbca3 100644
--- a/main.py
+++ b/main.py
@@ -1,546 +1,99 @@
"""
-Designed and Developed by-
-Udayraj Deshmukh
-https://github.com/Udayraj123
+ OMRChecker
+
+ Author: Udayraj Deshmukh
+ Github: https://github.com/Udayraj123
"""
-import re
-import os
-import cv2
import argparse
-import numpy as np
-import pandas as pd
-import matplotlib.pyplot as plt
-
-# from utils import * #Now imported via template
-from globals import *
-from template import *
-from glob import glob
-from csv import QUOTE_NONNUMERIC
-from time import localtime, strftime, time
-
-
-# TODO(beginner task) :-
-# from colorama import init
-# init()
-# from colorama import Fore, Back, Style
-
-def process_dir(root_dir, subdir, template):
- curr_dir = os.path.join(root_dir, subdir)
-
- # Look for template in current dir
- template_file = os.path.join(curr_dir, TEMPLATE_FILE)
- if os.path.exists(template_file):
- template = Template(template_file)
-
- # look for images in current dir to process
- paths = Paths(os.path.join(args['output_dir'], subdir))
- exts = ('*.png', '*.jpg')
- omr_files = sorted(
- [f for ext in exts for f in glob(os.path.join(curr_dir, ext))])
-
- # Exclude marker image if exists
- if(template and template.marker_path):
- omr_files = [f for f in omr_files if f != template.marker_path]
-
- subfolders = sorted([file for file in os.listdir(
- curr_dir) if os.path.isdir(os.path.join(curr_dir, file))])
- if omr_files:
- args_local = args.copy()
- if("OverrideFlags" in template.options):
- args_local.update(template.options["OverrideFlags"])
- print('\n------------------------------------------------------------------')
- print(f'Processing directory "{curr_dir}" with settings- ')
- print("\tTotal images : %d" % (len(omr_files)))
- print("\tCropping Enabled : " + str(not args_local["noCropping"]))
- print("\tAuto Alignment : " + str(args_local["autoAlign"]))
- print("\tUsing Template : " + str(template.path) if(template) else "N/A")
- print("\tUsing Marker : " + str(template.marker_path)
- if(template.marker is not None) else "N/A")
- print('')
-
- if not template:
- print(f'Error: No template file when processing {curr_dir}.')
- print(
- f' Place {TEMPLATE_FILE} in the directory or specify a template using -t.')
- return
-
- setup_dirs(paths)
- output_set = setup_output(paths, template)
- process_files(omr_files, template, args_local, output_set)
- elif(len(subfolders) == 0):
- # the directory should have images or be non-leaf
- print(f'Note: No valid images or subfolders found in {curr_dir}')
-
- # recursively process subfolders
- for folder in subfolders:
- process_dir(root_dir, os.path.join(subdir, folder), template)
-
-
-def checkAndMove(error_code, filepath, filepath2):
- # print("Dummy Move: "+filepath, " --> ",filepath2)
- global filesNotMoved
- filesNotMoved += 1
- return True
-
- global filesMoved
- if(not os.path.exists(filepath)):
- print('File already moved')
- return False
- if(os.path.exists(filepath2)):
- print('ERROR : Duplicate file at ' + filepath2)
- return False
-
- print("Moved: " + filepath, " --> ", filepath2)
- os.rename(filepath, filepath2)
- filesMoved += 1
- return True
-
-
-def processOMR(template, omrResp):
- # Note: This is a reference function. It is not part of the OMR checker
- # So its implementation is completely subjective to user's requirements.
- csvResp = {}
-
- # symbol for absent response
- UNMARKED_SYMBOL = ''
-
- # print("omrResp",omrResp)
-
- # Multi-column/multi-row questions which need to be concatenated
- for qNo, respKeys in template.concats.items():
- csvResp[qNo] = ''.join([omrResp.get(k, UNMARKED_SYMBOL)
- for k in respKeys])
-
- # Single-column/single-row questions
- for qNo in template.singles:
- csvResp[qNo] = omrResp.get(qNo, UNMARKED_SYMBOL)
-
- # Note: Concatenations and Singles together should be mutually exclusive
- # and should cover all questions in the template(exhaustive)
- # TODO: ^add a warning if omrResp has unused keys remaining
- return csvResp
-
-
-def report(
- Status,
- streak,
- scheme,
- qNo,
- marked,
- ans,
- prevmarks,
- currmarks,
- marks):
- print(
- '%s \t %s \t\t %s \t %s \t %s \t %s \t %s ' % (qNo,
- Status,
- str(streak),
- '[' + scheme + '] ',
- (str(prevmarks) + ' + ' + str(currmarks) + ' =' + str(marks)),
- str(marked),
- str(ans)))
-
-# check sectionwise only.
-
-
-def evaluate(resp, squad="H", explain=False):
- # TODO: @contributors - Need help generalizing this function
- global Answers, Sections
- marks = 0
- answers = Answers[squad]
- if(explain):
- print('Question\tStatus \t Streak\tSection \tMarks_Update\tMarked:\tAnswer:')
- for scheme, section in Sections[squad].items():
- sectionques = section['ques']
- prevcorrect = None
- allflag = 1
- streak = 0
- for q in sectionques:
- qNo = 'q' + str(q)
- ans = answers[qNo]
- marked = resp.get(qNo, 'X')
- firstQ = sectionques[0]
- lastQ = sectionques[len(sectionques) - 1]
- unmarked = marked == 'X' or marked == ''
- bonus = 'BONUS' in ans
- correct = bonus or (marked in ans)
- inrange = 0
-
- if(unmarked or int(q) == firstQ):
- streak = 0
- elif(prevcorrect == correct):
- streak += 1
- else:
- streak = 0
-
- if('allNone' in scheme):
- # loop on all sectionques
- allflag = allflag and correct
- if(q == lastQ):
- # at the end check allflag
- prevcorrect = correct
- currmarks = section['marks'] if allflag else 0
- else:
- currmarks = 0
-
- elif('Proxy' in scheme):
- a = int(ans[0])
- # proximity check
- inrange = 1 if unmarked else (
- float(abs(int(marked) - a)) / float(a) <= 0.25)
- currmarks = section['+marks'] if correct else (
- 0 if inrange else -section['-marks'])
-
- elif('Fibo' in scheme or 'Power' in scheme or 'Boom' in scheme):
- currmarks = section['+seq'][streak] if correct else (
- 0 if unmarked else -section['-seq'][streak])
- elif('TechnoFin' in scheme):
- currmarks = 0
- else:
- print('Invalid Sections')
- prevmarks = marks
- marks += currmarks
-
- if(explain):
- if bonus:
- report('BonusQ', streak, scheme, qNo, marked,
- ans, prevmarks, currmarks, marks)
- elif correct:
- report('Correct', streak, scheme, qNo, marked,
- ans, prevmarks, currmarks, marks)
- elif unmarked:
- report('Unmarked', streak, scheme, qNo, marked,
- ans, prevmarks, currmarks, marks)
- elif inrange:
- report('InProximity', streak, scheme, qNo,
- marked, ans, prevmarks, currmarks, marks)
- else:
- report('Incorrect', streak, scheme, qNo,
- marked, ans, prevmarks, currmarks, marks)
-
- prevcorrect = correct
-
- return marks
-
-
-def setup_output(paths, template):
- ns = argparse.Namespace()
- print("\nChecking Files...")
-
- # Include current output paths
- ns.paths = paths
-
- # custom sort: To use integer order in question names instead of
- # alphabetical - avoids q1, q10, q2 and orders them q1, q2, ..., q10
- ns.respCols = sorted(list(template.concats.keys()) + template.singles,
- key=lambda x: int(x[1:]) if ord(x[1]) in range(48, 58) else 0)
- ns.emptyResp = [''] * len(ns.respCols)
- ns.sheetCols = ['file_id', 'input_path',
- 'output_path', 'score'] + ns.respCols
- ns.OUTPUT_SET = []
- ns.filesObj = {}
- ns.filesMap = {
- "Results": paths.resultDir + 'Results_' + timeNowHrs + '.csv',
- "MultiMarked": paths.manualDir + 'MultiMarkedFiles_.csv',
- "Errors": paths.manualDir + 'ErrorFiles_.csv',
- "BadRollNos": paths.manualDir + 'BadRollNoFiles_.csv'
- }
-
- for fileKey, fileName in ns.filesMap.items():
- if(not os.path.exists(fileName)):
- print("Note: Created new file: %s" % (fileName))
- # still append mode req [THINK!]
- ns.filesObj[fileKey] = open(fileName, 'a')
- # Create Header Columns
- pd.DataFrame([ns.sheetCols], dtype=str).to_csv(
- ns.filesObj[fileKey], quoting=QUOTE_NONNUMERIC, header=False, index=False)
- else:
- print('Present : appending to %s' % (fileName))
- ns.filesObj[fileKey] = open(fileName, 'a')
-
- return ns
-
-
-''' TODO: Refactor into new process flow.
- Currently I have no idea what this does so I left it out'''
-
-
-def preliminary_check():
- filesCounter = 0
- # PRELIM_CHECKS for thresholding
- if(PRELIM_CHECKS):
- # TODO: add more using unit testing
- ALL_WHITE = 255 * \
- np.ones((TEMPLATE.dims[1], TEMPLATE.dims[0]), dtype='uint8')
- OMRresponseDict, final_marked, MultiMarked, multiroll = readResponse(
- ALL_WHITE, name="ALL_WHITE", savedir=None, autoAlign=True)
- print("ALL_WHITE", OMRresponseDict)
- if(OMRresponseDict != {}):
- print("Preliminary Checks Failed.")
- exit(12)
- ALL_BLACK = np.zeros(
- (TEMPLATE.dims[1], TEMPLATE.dims[0]), dtype='uint8')
- OMRresponseDict, final_marked, MultiMarked, multiroll = readResponse(
- ALL_BLACK, name="ALL_BLACK", savedir=None, autoAlign=True)
- print("ALL_BLACK", OMRresponseDict)
- show("Confirm : All bubbles are black", final_marked, 1, 1)
-
-
-def process_files(omr_files, template, args, out):
- start_time = int(time())
- filesCounter = 0
- filesNotMoved = 0
-
- for filepath in omr_files:
- filesCounter += 1
- # For windows filesystem support: all '\' will be replaced by '/'
- filepath = filepath.replace(os.sep, '/')
-
- # Prefixing a 'r' to use raw string (escape character '\' is taken
- # literally)
- finder = re.search(r'.*/(.*)/(.*)', filepath, re.IGNORECASE)
- if(finder):
- inputFolderName, filename = finder.groups()
- else:
- print("Error: Filepath not matching to Regex: " + filepath)
- continue
- # set global var for reading
-
- inOMR = cv2.imread(filepath, cv2.IMREAD_GRAYSCALE)
- print(
- '\n[%d] Processing image: \t' %
- (filesCounter),
- filepath,
- "\tResolution: ",
- inOMR.shape)
-
- OMRCrop = getROI(inOMR, filename, noCropping=args["noCropping"])
-
- if(OMRCrop is None):
- # Error OMR - could not crop
- newfilepath = out.paths.errorsDir + filename
- out.OUTPUT_SET.append([filename] + out.emptyResp)
- if(checkAndMove(NO_MARKER_ERR, filepath, newfilepath)):
- err_line = [filename, filepath,
- newfilepath, "NA"] + out.emptyResp
- pd.DataFrame(
- err_line,
- dtype=str).T.to_csv(
- out.filesObj["Errors"],
- quoting=QUOTE_NONNUMERIC,
- header=False,
- index=False)
- continue
-
- if template.marker is not None:
- OMRCrop = handle_markers(OMRCrop, template.marker, filename)
-
- if(args["setLayout"]):
- templateLayout = drawTemplateLayout(
- OMRCrop, template, shifted=False, border=2)
- show("Template Layout", templateLayout, 1, 1)
- continue
-
- # uniquify
- file_id = inputFolderName + '_' + filename
- savedir = out.paths.saveMarkedDir
- OMRresponseDict, final_marked, MultiMarked, multiroll = \
- readResponse(template, OMRCrop, name=file_id,
- savedir=savedir, autoAlign=args["autoAlign"])
-
- # concatenate roll nos, set unmarked responses, etc
- resp = processOMR(template, OMRresponseDict)
- print("\nRead Response: \t", resp)
-
- # This evaluates and returns the score attribute
- score = evaluate(resp, explain=explain)
- respArray = []
- for k in out.respCols:
- respArray.append(resp[k])
-
- out.OUTPUT_SET.append([filename] + respArray)
-
- # TODO: Add roll number validation here
- if(MultiMarked == 0):
- filesNotMoved += 1
- newfilepath = savedir + file_id
- # Enter into Results sheet-
- results_line = [filename, filepath, newfilepath, score] + respArray
- # Write/Append to results_line file(opened in append mode)
- pd.DataFrame(
- results_line,
- dtype=str).T.to_csv(
- out.filesObj["Results"],
- quoting=QUOTE_NONNUMERIC,
- header=False,
- index=False)
- print("[%d] Graded with score: %.2f" %
- (filesCounter, score), '\t file_id: ', file_id)
- # print(filesCounter,file_id,resp['Roll'],'score : ',score)
- else:
- # MultiMarked file
- print('[%d] MultiMarked, moving File: %s' %
- (filesCounter, file_id))
- newfilepath = out.paths.multiMarkedDir + filename
- if(checkAndMove(MULTI_BUBBLE_WARN, filepath, newfilepath)):
- mm_line = [filename, filepath, newfilepath, "NA"] + respArray
- pd.DataFrame(
- mm_line,
- dtype=str).T.to_csv(
- out.filesObj["MultiMarked"],
- quoting=QUOTE_NONNUMERIC,
- header=False,
- index=False)
- # else:
- # TODO: Add appropriate record handling here
- # pass
-
- # flush after every 20 files for a live view
- if(filesCounter % 20 == 0 or filesCounter == len(omr_files)):
- for fileKey in out.filesMap.keys():
- out.filesObj[fileKey].flush()
-
- timeChecking = round(time() - start_time, 2) if filesCounter else 1
- print('')
- print('Total files moved : %d ' % (filesMoved))
- print('Total files not moved : %d ' % (filesNotMoved))
- print('------------------------------')
- print(
- 'Total files processed : %d (%s)' %
- (filesCounter,
- 'Sum Tallied!' if filesCounter == (
- filesMoved +
- filesNotMoved) else 'Not Tallying!'))
-
- if(showimglvl <= 0):
- print(
- '\nFinished Checking %d files in %.1f seconds i.e. ~%.1f minutes.' %
- (filesCounter, timeChecking, timeChecking / 60))
- print('OMR Processing Rate :\t ~ %.2f seconds/OMR' %
- (timeChecking / filesCounter))
- print('OMR Processing Speed :\t ~ %.2f OMRs/minute' %
- ((filesCounter * 60) / timeChecking))
- else:
- print("\nTotal script time :", timeChecking, "seconds")
-
- if(showimglvl <= 1):
- # TODO: colorama this
- print(
- "\nTip: To see some awesome visuals, open globals.py and increase 'showimglvl'")
-
- evaluate_correctness(template, out)
-
- # Use this data to train as +ve feedback
- if(showimglvl >= 0 and filesCounter > 10):
- # TODO: Find good parameters to plot and depict image set quality
- for x in [thresholdCircles]: # ,badThresholds,veryBadPoints, , mbs]:
- if(x != []):
- x = pd.DataFrame(x)
- print(x.describe())
- plt.plot(range(len(x)), x)
- plt.title("Mystery Plot")
- plt.show()
- else:
- print(x)
-
-
-# Evaluate accuracy based on OMRDataset file generated through moderation
-# portal on the same set of images
-def evaluate_correctness(template, out):
- # TODO: TEST_FILE WOULD BE RELATIVE TO INPUT SUBDIRECTORY NOW-
- TEST_FILE = 'inputs/OMRDataset.csv'
- if(os.path.exists(TEST_FILE)):
- print("\nStarting evaluation for: " + TEST_FILE)
-
- TEST_COLS = ['file_id'] + out.respCols
- y_df = pd.read_csv(
- TEST_FILE, dtype=str)[TEST_COLS].replace(
- np.nan, '', regex=True).set_index('file_id')
-
- if(np.any(y_df.index.duplicated)):
- y_df_filtered = y_df.loc[~y_df.index.duplicated(keep='first')]
- print(
- "WARNING: Found duplicate File-ids in file %s. Removed %d rows from testing data. Rows remaining: %d" %
- (TEST_FILE, y_df.shape[0] - y_df_filtered.shape[0], y_df_filtered.shape[0]))
- y_df = y_df_filtered
-
- x_df = pd.DataFrame(
- out.OUTPUT_SET,
- dtype=str,
- columns=TEST_COLS).set_index('file_id')
- # print("x_df",x_df.head())
- # print("\ny_df",y_df.head())
- intersection = y_df.index.intersection(x_df.index)
-
- # Checking if the merge is okay
- if(intersection.size == x_df.index.size):
- y_df = y_df.loc[intersection]
- x_df['TestResult'] = (x_df == y_df).all(axis=1).astype(int)
- print(x_df.head())
- print("\n\t Accuracy on the %s Dataset: %.6f" %
- (TEST_FILE, (x_df['TestResult'].sum() / x_df.shape[0])))
- else:
- print(
- "\nERROR: Insufficient Testing Data: Have you appended MultiMarked data yet?")
- print("Missing File-ids: ",
- list(x_df.index.difference(intersection)))
-
-
-timeNowHrs = strftime("%I%p", localtime())
-
-# construct the argument parse and parse the arguments
-argparser = argparse.ArgumentParser()
-# https://docs.python.org/3/howto/argparse.html
-# store_true: if the option is specified, assign the value True to
-# args.verbose. Not specifying it implies False.
-argparser.add_argument(
- "-c",
- "--noCropping",
- required=False,
- dest='noCropping',
- action='store_true',
- help="Disables page contour detection - used when page boundary is not visible e.g. document scanner.")
-argparser.add_argument(
- "-a",
- "--autoAlign",
- required=False,
- dest='autoAlign',
- action='store_true',
- help="(experimental) Enables automatic template alignment - use if the scans show slight misalignments.")
-argparser.add_argument(
- "-l",
- "--setLayout",
- required=False,
- dest='setLayout',
- action='store_true',
- help="Set up OMR template layout - modify your json file and run again until the template is set.")
-argparser.add_argument("-i", "--inputDir", required=False, action='append',
- dest='input_dir', help="Specify an input directory.")
-argparser.add_argument("-o", "--outputDir", default='outputs', required=False,
- dest='output_dir', help="Specify an output directory.")
-argparser.add_argument(
- "-t",
- "--template",
- required=False,
- dest='template',
- help="Specify a default template if no template file in input directories.")
-
-
-args, unknown = argparser.parse_known_args()
-args = vars(args)
-if(len(unknown) > 0):
- print("\nError: Unknown arguments:", unknown)
- argparser.print_help()
- exit(11)
-
-if args['template']:
- args['template'] = Template(args['template'])
-
-if args['input_dir'] is None:
- args['input_dir'] = ['inputs']
-
-for root in args['input_dir']:
- process_dir(root, '', args['template'])
+import sys
+from pathlib import Path
+
+from src.entry import entry_point
+from src.logger import logger
+
+
+def parse_args():
+ # construct the argument parse and parse the arguments
+ argparser = argparse.ArgumentParser()
+
+ argparser.add_argument(
+ "-i",
+ "--inputDir",
+ default=["inputs"],
+ # https://docs.python.org/3/library/argparse.html#nargs
+ nargs="*",
+ required=False,
+ type=str,
+ dest="input_paths",
+ help="Specify an input directory.",
+ )
+
+ argparser.add_argument(
+ "-d",
+ "--debug",
+ required=False,
+ dest="debug",
+ action="store_false",
+ help="Enables debugging mode for showing detailed errors",
+ )
+
+ argparser.add_argument(
+ "-o",
+ "--outputDir",
+ default="outputs",
+ required=False,
+ dest="output_dir",
+ help="Specify an output directory.",
+ )
+
+ argparser.add_argument(
+ "-a",
+ "--autoAlign",
+ required=False,
+ dest="autoAlign",
+ action="store_true",
+ help="(experimental) Enables automatic template alignment - \
+ use if the scans show slight misalignments.",
+ )
+
+ argparser.add_argument(
+ "-l",
+ "--setLayout",
+ required=False,
+ dest="setLayout",
+ action="store_true",
+ help="Set up OMR template layout - modify your json file and \
+ run again until the template is set.",
+ )
+
+ (
+ args,
+ unknown,
+ ) = argparser.parse_known_args()
+
+ args = vars(args)
+
+ if len(unknown) > 0:
+ logger.warning(f"\nError: Unknown arguments: {unknown}", unknown)
+ argparser.print_help()
+ exit(11)
+ return args
+
+
+def entry_point_for_args(args):
+ if args["debug"] is True:
+ # Disable tracebacks
+ sys.tracebacklimit = 0
+ for root in args["input_paths"]:
+ entry_point(
+ Path(root),
+ args,
+ )
+
+
+if __name__ == "__main__":
+ args = parse_args()
+ entry_point_for_args(args)
diff --git a/pyproject.toml b/pyproject.toml
new file mode 100644
index 00000000..060309be
--- /dev/null
+++ b/pyproject.toml
@@ -0,0 +1,18 @@
+[tool.black]
+exclude = '''
+(
+ /(
+ \.eggs # exclude a few common directories in the
+ | \.git # root of the project
+ | \.venv
+ | _build
+ | build
+ | dist
+ )/
+ | foo.py # also separately exclude a file named foo.py in
+ # the root of the project
+)
+'''
+include = '\.pyi?$'
+line-length = 88
+target-version = ['py37']
diff --git a/pytest.ini b/pytest.ini
new file mode 100644
index 00000000..84008a21
--- /dev/null
+++ b/pytest.ini
@@ -0,0 +1,6 @@
+# pytest.ini
+[pytest]
+minversion = 7.0
+addopts = -qq --capture=no
+testpaths =
+ src/tests
diff --git a/requirements.dev.txt b/requirements.dev.txt
new file mode 100644
index 00000000..722ab0dd
--- /dev/null
+++ b/requirements.dev.txt
@@ -0,0 +1,7 @@
+-r requirements.txt
+flake8>=6.0.0
+freezegun>=1.2.2
+pre-commit>=3.3.3
+pytest-mock>=3.11.1
+pytest>=7.4.0
+syrupy>=4.0.4
diff --git a/requirements.txt b/requirements.txt
index 505865c9..761d2e51 100644
--- a/requirements.txt
+++ b/requirements.txt
@@ -1,4 +1,8 @@
-imutils>=0.5.2
-matplotlib>=3.0.2
-numpy>=1.16.0
-pandas>=0.24.0
+deepmerge>=1.1.0
+dotmap>=1.3.30
+jsonschema>=4.17.3
+matplotlib>=3.7.1
+numpy>=1.25.0
+pandas>=2.0.2
+rich>=13.4.2
+screeninfo>=0.8.1
diff --git a/samples/answer-key/using-csv/adrian_omr.png b/samples/answer-key/using-csv/adrian_omr.png
new file mode 100644
index 00000000..d8db0994
Binary files /dev/null and b/samples/answer-key/using-csv/adrian_omr.png differ
diff --git a/samples/answer-key/using-csv/answer_key.csv b/samples/answer-key/using-csv/answer_key.csv
new file mode 100644
index 00000000..566201d1
--- /dev/null
+++ b/samples/answer-key/using-csv/answer_key.csv
@@ -0,0 +1,5 @@
+q1,C
+q2,E
+q3,A
+q4,B
+q5,B
\ No newline at end of file
diff --git a/samples/answer-key/using-csv/evaluation.json b/samples/answer-key/using-csv/evaluation.json
new file mode 100644
index 00000000..14a3db25
--- /dev/null
+++ b/samples/answer-key/using-csv/evaluation.json
@@ -0,0 +1,14 @@
+{
+ "source_type": "csv",
+ "options": {
+ "answer_key_csv_path": "answer_key.csv",
+ "should_explain_scoring": true
+ },
+ "marking_schemes": {
+ "DEFAULT": {
+ "correct": "1",
+ "incorrect": "0",
+ "unmarked": "0"
+ }
+ }
+}
diff --git a/samples/answer-key/using-csv/template.json b/samples/answer-key/using-csv/template.json
new file mode 100644
index 00000000..41ec9ffa
--- /dev/null
+++ b/samples/answer-key/using-csv/template.json
@@ -0,0 +1,35 @@
+{
+ "pageDimensions": [
+ 300,
+ 400
+ ],
+ "bubbleDimensions": [
+ 25,
+ 25
+ ],
+ "preProcessors": [
+ {
+ "name": "CropPage",
+ "options": {
+ "morphKernel": [
+ 10,
+ 10
+ ]
+ }
+ }
+ ],
+ "fieldBlocks": {
+ "MCQ_Block_1": {
+ "fieldType": "QTYPE_MCQ5",
+ "origin": [
+ 65,
+ 60
+ ],
+ "fieldLabels": [
+ "q1..5"
+ ],
+ "labelsGap": 52,
+ "bubblesGap": 41
+ }
+ }
+}
diff --git a/samples/answer-key/weighted-answers/evaluation.json b/samples/answer-key/weighted-answers/evaluation.json
new file mode 100644
index 00000000..c0daefcf
--- /dev/null
+++ b/samples/answer-key/weighted-answers/evaluation.json
@@ -0,0 +1,35 @@
+{
+ "source_type": "custom",
+ "options": {
+ "questions_in_order": [
+ "q1..5"
+ ],
+ "answers_in_order": [
+ "C",
+ "E",
+ [
+ "A",
+ "C"
+ ],
+ [
+ [
+ "B",
+ 2
+ ],
+ [
+ "C",
+ "3/2"
+ ]
+ ],
+ "C"
+ ],
+ "should_explain_scoring": true
+ },
+ "marking_schemes": {
+ "DEFAULT": {
+ "correct": "3",
+ "incorrect": "-1",
+ "unmarked": "0"
+ }
+ }
+}
diff --git a/samples/answer-key/weighted-answers/images/adrian_omr.png b/samples/answer-key/weighted-answers/images/adrian_omr.png
new file mode 100644
index 00000000..69a53823
Binary files /dev/null and b/samples/answer-key/weighted-answers/images/adrian_omr.png differ
diff --git a/samples/answer-key/weighted-answers/images/adrian_omr_2.png b/samples/answer-key/weighted-answers/images/adrian_omr_2.png
new file mode 100644
index 00000000..d8db0994
Binary files /dev/null and b/samples/answer-key/weighted-answers/images/adrian_omr_2.png differ
diff --git a/samples/answer-key/weighted-answers/template.json b/samples/answer-key/weighted-answers/template.json
new file mode 100644
index 00000000..41ec9ffa
--- /dev/null
+++ b/samples/answer-key/weighted-answers/template.json
@@ -0,0 +1,35 @@
+{
+ "pageDimensions": [
+ 300,
+ 400
+ ],
+ "bubbleDimensions": [
+ 25,
+ 25
+ ],
+ "preProcessors": [
+ {
+ "name": "CropPage",
+ "options": {
+ "morphKernel": [
+ 10,
+ 10
+ ]
+ }
+ }
+ ],
+ "fieldBlocks": {
+ "MCQ_Block_1": {
+ "fieldType": "QTYPE_MCQ5",
+ "origin": [
+ 65,
+ 60
+ ],
+ "fieldLabels": [
+ "q1..5"
+ ],
+ "labelsGap": 52,
+ "bubblesGap": 41
+ }
+ }
+}
diff --git a/samples/community/Antibodyy/simple_omr_sheet.jpg b/samples/community/Antibodyy/simple_omr_sheet.jpg
new file mode 100644
index 00000000..661d5f4f
Binary files /dev/null and b/samples/community/Antibodyy/simple_omr_sheet.jpg differ
diff --git a/samples/community/Antibodyy/template.json b/samples/community/Antibodyy/template.json
new file mode 100644
index 00000000..7e962bbf
--- /dev/null
+++ b/samples/community/Antibodyy/template.json
@@ -0,0 +1,35 @@
+{
+ "pageDimensions": [
+ 299,
+ 398
+ ],
+ "bubbleDimensions": [
+ 42,
+ 42
+ ],
+ "fieldBlocks": {
+ "MCQBlock1": {
+ "fieldType": "QTYPE_MCQ5",
+ "origin": [
+ 65,
+ 79
+ ],
+ "bubblesGap": 43,
+ "labelsGap": 50,
+ "fieldLabels": [
+ "q1..6"
+ ]
+ }
+ },
+ "preProcessors": [
+ {
+ "name": "CropPage",
+ "options": {
+ "morphKernel": [
+ 10,
+ 10
+ ]
+ }
+ }
+ ]
+}
diff --git a/samples/community/Sandeep-1507/omr-1.png b/samples/community/Sandeep-1507/omr-1.png
new file mode 100644
index 00000000..8c40655e
Binary files /dev/null and b/samples/community/Sandeep-1507/omr-1.png differ
diff --git a/samples/community/Sandeep-1507/omr-2.png b/samples/community/Sandeep-1507/omr-2.png
new file mode 100644
index 00000000..aabef0a5
Binary files /dev/null and b/samples/community/Sandeep-1507/omr-2.png differ
diff --git a/samples/community/Sandeep-1507/omr-3.png b/samples/community/Sandeep-1507/omr-3.png
new file mode 100644
index 00000000..8f1cb5c8
Binary files /dev/null and b/samples/community/Sandeep-1507/omr-3.png differ
diff --git a/samples/community/Sandeep-1507/template.json b/samples/community/Sandeep-1507/template.json
new file mode 100644
index 00000000..b35404e2
--- /dev/null
+++ b/samples/community/Sandeep-1507/template.json
@@ -0,0 +1,234 @@
+{
+ "pageDimensions": [
+ 1189,
+ 1682
+ ],
+ "bubbleDimensions": [
+ 15,
+ 15
+ ],
+ "preProcessors": [
+ {
+ "name": "GaussianBlur",
+ "options": {
+ "kSize": [
+ 3,
+ 3
+ ],
+ "sigmaX": 0
+ }
+ }
+ ],
+ "customLabels": {
+ "Booklet_No": [
+ "b1..7"
+ ]
+ },
+ "fieldBlocks": {
+ "Booklet_No": {
+ "fieldType": "QTYPE_INT",
+ "origin": [
+ 112,
+ 530
+ ],
+ "fieldLabels": [
+ "b1..7"
+ ],
+ "emptyValue": "no",
+ "bubblesGap": 28,
+ "labelsGap": 26.5
+ },
+ "MCQBlock1a1": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q1..10"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 476,
+ 100
+ ]
+ },
+ "MCQBlock1a2": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q11..20"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 476,
+ 370
+ ]
+ },
+ "MCQBlock1a3": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q21..35"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 476,
+ 638
+ ]
+ },
+ "MCQBlock2a1": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q51..60"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 645,
+ 100
+ ]
+ },
+ "MCQBlock2a2": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q61..70"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 645,
+ 370
+ ]
+ },
+ "MCQBlock2a3": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q71..85"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 645,
+ 638
+ ]
+ },
+ "MCQBlock3a1": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q101..110"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 815,
+ 100
+ ]
+ },
+ "MCQBlock3a2": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q111..120"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 815,
+ 370
+ ]
+ },
+ "MCQBlock3a3": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q121..135"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 815,
+ 638
+ ]
+ },
+ "MCQBlock4a1": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q151..160"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 983,
+ 100
+ ]
+ },
+ "MCQBlock4a2": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q161..170"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 983,
+ 370
+ ]
+ },
+ "MCQBlock4a3": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q171..185"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 983,
+ 638
+ ]
+ },
+ "MCQBlock1a": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q36..50"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 480,
+ 1061
+ ]
+ },
+ "MCQBlock2a": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q86..100"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 648,
+ 1061
+ ]
+ },
+ "MCQBlock3a": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q136..150"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.7,
+ "origin": [
+ 815,
+ 1061
+ ]
+ },
+ "MCQBlock4a": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q186..200"
+ ],
+ "bubblesGap": 28.7,
+ "labelsGap": 26.6,
+ "origin": [
+ 986,
+ 1061
+ ]
+ }
+ }
+}
diff --git a/samples/community/Shamanth/omr_sheet_01.png b/samples/community/Shamanth/omr_sheet_01.png
new file mode 100644
index 00000000..ead17dd1
Binary files /dev/null and b/samples/community/Shamanth/omr_sheet_01.png differ
diff --git a/samples/community/Shamanth/template.json b/samples/community/Shamanth/template.json
new file mode 100644
index 00000000..fb18d975
--- /dev/null
+++ b/samples/community/Shamanth/template.json
@@ -0,0 +1,25 @@
+{
+ "pageDimensions": [
+ 300,
+ 400
+ ],
+ "bubbleDimensions": [
+ 20,
+ 20
+ ],
+ "fieldBlocks": {
+ "MCQBlock1": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 78,
+ 41
+ ],
+ "fieldLabels": [
+ "q21..28"
+ ],
+ "bubblesGap": 56,
+ "labelsGap": 46
+ }
+ },
+ "preProcessors": []
+}
diff --git a/samples/community/UPSC-mock/answer_key.jpg b/samples/community/UPSC-mock/answer_key.jpg
new file mode 100644
index 00000000..7a229c1b
Binary files /dev/null and b/samples/community/UPSC-mock/answer_key.jpg differ
diff --git a/samples/community/UPSC-mock/config.json b/samples/community/UPSC-mock/config.json
new file mode 100644
index 00000000..0edf9d22
--- /dev/null
+++ b/samples/community/UPSC-mock/config.json
@@ -0,0 +1,11 @@
+{
+ "dimensions": {
+ "display_height": 1800,
+ "display_width": 2400,
+ "processing_height": 2400,
+ "processing_width": 1800
+ },
+ "outputs": {
+ "show_image_level": 0
+ }
+}
diff --git a/samples/community/UPSC-mock/evaluation.json b/samples/community/UPSC-mock/evaluation.json
new file mode 100644
index 00000000..42fbec8b
--- /dev/null
+++ b/samples/community/UPSC-mock/evaluation.json
@@ -0,0 +1,18 @@
+{
+ "source_type": "csv",
+ "options": {
+ "answer_key_csv_path": "answer_key.csv",
+ "answer_key_image_path": "answer_key.jpg",
+ "questions_in_order": [
+ "q1..100"
+ ],
+ "should_explain_scoring": true
+ },
+ "marking_schemes": {
+ "DEFAULT": {
+ "correct": "2",
+ "incorrect": "-2/3",
+ "unmarked": "0"
+ }
+ }
+}
diff --git a/samples/community/UPSC-mock/scan-angles/angle-1.jpg b/samples/community/UPSC-mock/scan-angles/angle-1.jpg
new file mode 100644
index 00000000..9c75d7be
Binary files /dev/null and b/samples/community/UPSC-mock/scan-angles/angle-1.jpg differ
diff --git a/samples/community/UPSC-mock/scan-angles/angle-2.jpg b/samples/community/UPSC-mock/scan-angles/angle-2.jpg
new file mode 100644
index 00000000..edab6060
Binary files /dev/null and b/samples/community/UPSC-mock/scan-angles/angle-2.jpg differ
diff --git a/samples/community/UPSC-mock/scan-angles/angle-3.jpg b/samples/community/UPSC-mock/scan-angles/angle-3.jpg
new file mode 100644
index 00000000..f1da4af9
Binary files /dev/null and b/samples/community/UPSC-mock/scan-angles/angle-3.jpg differ
diff --git a/samples/community/UPSC-mock/template.json b/samples/community/UPSC-mock/template.json
new file mode 100644
index 00000000..a1ef5c6b
--- /dev/null
+++ b/samples/community/UPSC-mock/template.json
@@ -0,0 +1,195 @@
+{
+ "pageDimensions": [
+ 1800,
+ 2400
+ ],
+ "bubbleDimensions": [
+ 30,
+ 25
+ ],
+ "customLabels": {
+ "Subject Code": [
+ "subjectCode1",
+ "subjectCode2"
+ ],
+ "Roll": [
+ "roll1..10"
+ ]
+ },
+ "fieldBlocks": {
+ "bookletNo": {
+ "origin": [
+ 595,
+ 545
+ ],
+ "bubblesGap": 68,
+ "labelsGap": 0,
+ "fieldLabels": [
+ "bookletNo"
+ ],
+ "bubbleValues": [
+ "A",
+ "B",
+ "C",
+ "D"
+ ],
+ "direction": "vertical"
+ },
+ "subjectCode": {
+ "origin": [
+ 912,
+ 512
+ ],
+ "bubblesGap": 33,
+ "labelsGap": 42.5,
+ "fieldLabels": [
+ "subjectCode1",
+ "subjectCode2"
+ ],
+ "fieldType": "QTYPE_INT"
+ },
+ "roll": {
+ "origin": [
+ 1200,
+ 510
+ ],
+ "bubblesGap": 33,
+ "labelsGap": 42.8,
+ "fieldLabels": [
+ "roll1..10"
+ ],
+ "fieldType": "QTYPE_INT"
+ },
+ "q01block": {
+ "origin": [
+ 500,
+ 927
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q1..10"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q11block": {
+ "origin": [
+ 500,
+ 1258
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q11..20"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q21block": {
+ "origin": [
+ 500,
+ 1589
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q21..30"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q31block": {
+ "origin": [
+ 495,
+ 1925
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q31..40"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q41block": {
+ "origin": [
+ 811,
+ 927
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q41..50"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q51block": {
+ "origin": [
+ 811,
+ 1258
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q51..60"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q61block": {
+ "origin": [
+ 811,
+ 1589
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q61..70"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q71block": {
+ "origin": [
+ 811,
+ 1925
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q71..80"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q81block": {
+ "origin": [
+ 1125,
+ 927
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q81..90"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q91block": {
+ "origin": [
+ 1125,
+ 1258
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q91..100"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ }
+ },
+ "preProcessors": [
+ {
+ "name": "CropPage",
+ "options": {
+ "morphKernel": [
+ 10,
+ 10
+ ]
+ }
+ }
+ ]
+}
diff --git a/samples/community/UmarFarootAPS/answer_key.csv b/samples/community/UmarFarootAPS/answer_key.csv
new file mode 100644
index 00000000..b40e8959
--- /dev/null
+++ b/samples/community/UmarFarootAPS/answer_key.csv
@@ -0,0 +1,200 @@
+q1,C
+q2,C
+q3,"D,E"
+q4,"A,AB"
+q5,"[['A', '1'], ['B', '2']]"
+q6,"['A', 'B']"
+q7,C
+q8,D
+q9,B
+q10,B
+q11,A
+q12,A
+q13,C
+q14,B
+q15,D
+q16,B
+q17,C
+q18,A
+q19,B
+q20,D
+q21,D
+q22,C
+q23,A
+q24,C
+q25,D
+q26,C
+q27,C
+q28,B
+q29,A
+q30,D
+q31,C
+q32,B
+q33,B
+q34,C
+q35,A
+q36,D
+q37,C
+q38,B
+q39,C
+q40,A
+q41,A
+q42,C
+q43,D
+q44,D
+q45,B
+q46,C
+q47,C
+q48,A
+q49,C
+q50,B
+q51,B
+q52,C
+q53,D
+q54,C
+q55,B
+q56,B
+q57,A
+q58,A
+q59,D
+q60,C
+q61,C
+q62,B
+q63,A
+q64,C
+q65,D
+q66,C
+q67,B
+q68,A
+q69,B
+q70,B
+q71,C
+q72,B
+q73,C
+q74,A
+q75,A
+q76,C
+q77,D
+q78,D
+q79,B
+q80,A
+q81,B
+q82,C
+q83,D
+q84,C
+q85,A
+q86,C
+q87,D
+q88,B
+q89,C
+q90,B
+q91,B
+q92,A
+q93,C
+q94,D
+q95,C
+q96,B
+q97,B
+q98,A
+q99,A
+q100,A
+q101,A
+q102,B
+q103,C
+q104,C
+q105,A
+q106,D
+q107,B
+q108,A
+q109,C
+q110,B
+q111,B
+q112,C
+q113,C
+q114,B
+q115,D
+q116,B
+q117,A
+q118,C
+q119,D
+q120,C
+q121,C
+q122,A
+q123,B
+q124,C
+q125,D
+q126,C
+q127,C
+q128,D
+q129,D
+q130,A
+q131,A
+q132,C
+q133,B
+q134,C
+q135,D
+q136,B
+q137,C
+q138,A
+q139,B
+q140,D
+q141,D
+q142,C
+q143,D
+q144,A
+q145,A
+q146,C
+q147,A
+q148,D
+q149,D
+q150,B
+q151,A
+q152,B
+q153,B
+q154,D
+q155,D
+q156,B
+q157,A
+q158,B
+q159,A
+q160,C
+q161,D
+q162,C
+q163,A
+q164,B
+q165,D
+q166,D
+q167,C
+q168,C
+q169,C
+q170,D
+q171,A
+q172,A
+q173,C
+q174,C
+q175,B
+q176,D
+q177,A
+q178,B
+q179,B
+q180,C
+q181,D
+q182,C
+q183,B
+q184,B
+q185,C
+q186,D
+q187,D
+q188,A
+q189,A
+q190,B
+q191,C
+q192,B
+q193,D
+q194,C
+q195,B
+q196,B
+q197,A
+q198,B
+q199,B
+q200,A
diff --git a/samples/community/UmarFarootAPS/config.json b/samples/community/UmarFarootAPS/config.json
new file mode 100644
index 00000000..fde8f5b2
--- /dev/null
+++ b/samples/community/UmarFarootAPS/config.json
@@ -0,0 +1,11 @@
+{
+ "dimensions": {
+ "display_height": 960,
+ "display_width": 1280,
+ "processing_height": 1640,
+ "processing_width": 1332
+ },
+ "outputs": {
+ "show_image_level": 0
+ }
+}
diff --git a/samples/community/UmarFarootAPS/evaluation.json b/samples/community/UmarFarootAPS/evaluation.json
new file mode 100644
index 00000000..14a3db25
--- /dev/null
+++ b/samples/community/UmarFarootAPS/evaluation.json
@@ -0,0 +1,14 @@
+{
+ "source_type": "csv",
+ "options": {
+ "answer_key_csv_path": "answer_key.csv",
+ "should_explain_scoring": true
+ },
+ "marking_schemes": {
+ "DEFAULT": {
+ "correct": "1",
+ "incorrect": "0",
+ "unmarked": "0"
+ }
+ }
+}
diff --git a/samples/sample3/omr_marker.jpg b/samples/community/UmarFarootAPS/omr_marker.jpg
similarity index 100%
rename from samples/sample3/omr_marker.jpg
rename to samples/community/UmarFarootAPS/omr_marker.jpg
diff --git a/samples/community/UmarFarootAPS/scans/scan-type-1.jpg b/samples/community/UmarFarootAPS/scans/scan-type-1.jpg
new file mode 100644
index 00000000..0932b240
Binary files /dev/null and b/samples/community/UmarFarootAPS/scans/scan-type-1.jpg differ
diff --git a/samples/community/UmarFarootAPS/scans/scan-type-2.jpg b/samples/community/UmarFarootAPS/scans/scan-type-2.jpg
new file mode 100644
index 00000000..2eef7c32
Binary files /dev/null and b/samples/community/UmarFarootAPS/scans/scan-type-2.jpg differ
diff --git a/samples/community/UmarFarootAPS/template.json b/samples/community/UmarFarootAPS/template.json
new file mode 100644
index 00000000..f096f518
--- /dev/null
+++ b/samples/community/UmarFarootAPS/template.json
@@ -0,0 +1,188 @@
+{
+ "pageDimensions": [
+ 2550,
+ 3300
+ ],
+ "bubbleDimensions": [
+ 32,
+ 32
+ ],
+ "preProcessors": [
+ {
+ "name": "CropOnMarkers",
+ "options": {
+ "relativePath": "omr_marker.jpg",
+ "sheetToMarkerWidthRatio": 17
+ }
+ }
+ ],
+ "customLabels": {
+ "Roll_no": [
+ "r1",
+ "r2",
+ "r3",
+ "r4"
+ ]
+ },
+ "fieldBlocks": {
+ "Roll_no": {
+ "fieldType": "QTYPE_INT",
+ "origin": [
+ 2169,
+ 180
+ ],
+ "fieldLabels": [
+ "r1",
+ "r2",
+ "r3",
+ "r4"
+ ],
+ "bubblesGap": 61,
+ "labelsGap": 93
+ },
+ "MCQBlock1a1": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 197,
+ 300
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q1..17"
+ ]
+ },
+ "MCQBlock1a2": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 197,
+ 1310
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q18..34"
+ ]
+ },
+ "MCQBlock1a3": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 197,
+ 2316
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q35..50"
+ ]
+ },
+ "MCQBlock1a4": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 725,
+ 300
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q51..67"
+ ]
+ },
+ "MCQBlock1a5": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 725,
+ 1310
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q68..84"
+ ]
+ },
+ "MCQBlock1a6": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 725,
+ 2316
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q85..100"
+ ]
+ },
+ "MCQBlock1a7": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 1250,
+ 300
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q101..117"
+ ]
+ },
+ "MCQBlock1a8": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 1250,
+ 1310
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q118..134"
+ ]
+ },
+ "MCQBlock1a9": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 1250,
+ 2316
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q135..150"
+ ]
+ },
+ "MCQBlock1a10": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 1770,
+ 300
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q151..167"
+ ]
+ },
+ "MCQBlock1a11": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 1770,
+ 1310
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q168..184"
+ ]
+ },
+ "MCQBlock1a12": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 1770,
+ 2316
+ ],
+ "bubblesGap": 92,
+ "labelsGap": 59.6,
+ "fieldLabels": [
+ "q185..200"
+ ]
+ }
+ }
+}
diff --git a/samples/community/dxuian/omrcollegesheet.jpg b/samples/community/dxuian/omrcollegesheet.jpg
new file mode 100644
index 00000000..b9d70c6b
Binary files /dev/null and b/samples/community/dxuian/omrcollegesheet.jpg differ
diff --git a/samples/community/dxuian/template.json b/samples/community/dxuian/template.json
new file mode 100644
index 00000000..43bf2be6
--- /dev/null
+++ b/samples/community/dxuian/template.json
@@ -0,0 +1,48 @@
+{
+ "pageDimensions": [707, 484],
+ "bubbleDimensions": [15, 10],
+ "fieldBlocks": {
+ "Column1": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [82, 35],
+ "bubblesGap": 21,
+ "labelsGap": 22.7,
+ "bubbleCount": 20,
+ "fieldLabels": ["Q1", "Q2", "Q3", "Q4", "Q5", "Q6", "Q7", "Q8", "Q9", "Q10", "Q11", "Q12", "Q13", "Q14", "Q15", "Q16", "Q17", "Q18", "Q19", "Q20"]
+ },
+ "Column2": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [205, 35],
+ "bubblesGap": 21,
+ "labelsGap": 22.7,
+ "bubbleCount": 20,
+ "fieldLabels": ["Q21", "Q22", "Q23", "Q24", "Q25", "Q26", "Q27", "Q28", "Q29", "Q30", "Q31", "Q32", "Q33", "Q34", "Q35", "Q36", "Q37", "Q38", "Q39", "Q40"]
+ },
+ "Column3": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [327, 35],
+ "bubblesGap": 21,
+ "labelsGap": 22.7,
+ "bubbleCount": 20,
+ "fieldLabels": ["Q41", "Q42", "Q43", "Q44", "Q45", "Q46", "Q47", "Q48", "Q49", "Q50", "Q51", "Q52", "Q53", "Q54", "Q55", "Q56", "Q57", "Q58", "Q59", "Q60"]
+ },
+ "Column4": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [450, 35],
+ "bubblesGap": 21,
+ "labelsGap": 22.7,
+ "bubbleCount": 20,
+ "fieldLabels": ["Q61", "Q62", "Q63", "Q64", "Q65", "Q66", "Q67", "Q68", "Q69", "Q70", "Q71", "Q72", "Q73", "Q74", "Q75", "Q76", "Q77", "Q78", "Q79", "Q80"]
+ },
+ "Column5": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [573, 35],
+ "bubblesGap": 21,
+ "labelsGap": 22.7,
+ "bubbleCount": 20,
+ "fieldLabels": ["Q81", "Q82", "Q83", "Q84", "Q85", "Q86", "Q87", "Q88", "Q89", "Q90", "Q91", "Q92", "Q93", "Q94", "Q95", "Q96", "Q97", "Q98", "Q99", "Q100"]
+ }
+ },
+
+ "emptyValue": "-"
+}
\ No newline at end of file
diff --git a/samples/community/ibrahimkilic/template.json b/samples/community/ibrahimkilic/template.json
new file mode 100644
index 00000000..f0cb9cf3
--- /dev/null
+++ b/samples/community/ibrahimkilic/template.json
@@ -0,0 +1,30 @@
+{
+ "pageDimensions": [
+ 299,
+ 328
+ ],
+ "bubbleDimensions": [
+ 20,
+ 20
+ ],
+ "emptyValue": "no",
+ "fieldBlocks": {
+ "YesNoBlock1": {
+ "direction": "horizontal",
+ "bubbleValues": [
+ "yes"
+ ],
+ "origin": [
+ 15,
+ 55
+ ],
+ "emptyValue": "no",
+ "bubblesGap": 48,
+ "labelsGap": 48,
+ "fieldLabels": [
+ "q1..5"
+ ]
+ }
+ },
+ "preProcessors": []
+}
diff --git a/samples/community/ibrahimkilic/yes_no_questionnarie.jpg b/samples/community/ibrahimkilic/yes_no_questionnarie.jpg
new file mode 100644
index 00000000..e9436f41
Binary files /dev/null and b/samples/community/ibrahimkilic/yes_no_questionnarie.jpg differ
diff --git a/samples/community/samuelIkoli/template.json b/samples/community/samuelIkoli/template.json
new file mode 100644
index 00000000..759f6972
--- /dev/null
+++ b/samples/community/samuelIkoli/template.json
@@ -0,0 +1,28 @@
+{
+ "pageDimensions": [630, 404],
+ "bubbleDimensions": [20, 15],
+ "customLabels": {},
+ "fieldBlocks": {
+ "MCQBlock1": {
+ "fieldType": "QTYPE_MCQ5",
+ "origin": [33, 6],
+ "fieldLabels": ["q1", "q2", "q3", "q4", "q5", "q6", "q7", "q8", "q9", "q10", "q11", "q12", "q13", "q14", "q15", "q16", "q17", "q18", "q19", "q20"],
+ "bubblesGap": 33,
+ "labelsGap": 20
+ },
+ "MCQBlock2": {
+ "fieldType": "QTYPE_MCQ5",
+ "origin": [248, 6],
+ "fieldLabels": ["q21", "q22", "q23", "q24", "q25", "q26", "q27", "q28", "q29", "q30", "q31", "q32", "q33", "q34", "q35", "q36", "q37", "q38", "q39", "q40"],
+ "bubblesGap": 33,
+ "labelsGap": 20
+ },
+ "MCQBlock3": {
+ "fieldType": "QTYPE_MCQ5",
+ "origin": [465, 6],
+ "fieldLabels": ["q41", "q42", "q43", "q44", "q45", "q46", "q47", "q48", "q49", "q50", "q51", "q52", "q53", "q54", "q55", "q56", "q57", "q58", "q59", "q60"],
+ "bubblesGap": 33,
+ "labelsGap": 20
+ }
+ }
+}
diff --git a/samples/community/samuelIkoli/waec_sample.jpeg b/samples/community/samuelIkoli/waec_sample.jpeg
new file mode 100644
index 00000000..fcd61e80
Binary files /dev/null and b/samples/community/samuelIkoli/waec_sample.jpeg differ
diff --git a/samples/sample1/config.json b/samples/sample1/config.json
new file mode 100644
index 00000000..a9daf449
--- /dev/null
+++ b/samples/sample1/config.json
@@ -0,0 +1,11 @@
+{
+ "dimensions": {
+ "display_height": 2480,
+ "display_width": 1640,
+ "processing_height": 820,
+ "processing_width": 666
+ },
+ "outputs": {
+ "show_image_level": 5
+ }
+}
diff --git a/samples/sample1/template.json b/samples/sample1/template.json
index ec0f9c68..46635774 100644
--- a/samples/sample1/template.json
+++ b/samples/sample1/template.json
@@ -1,260 +1,197 @@
{
- "Dimensions": [
+ "pageDimensions": [
1846,
1500
],
- "BubbleDimensions": [
+ "bubbleDimensions": [
40,
40
],
- "Options": {
- "Marker": {
- "RelativePath": "omr_marker.jpg",
- "SheetToMarkerWidthRatio": 17
- }
- },
- "Concatenations": {
+ "customLabels": {
"Roll": [
- "Squad",
"Medium",
- "roll0",
- "roll1",
- "roll2",
- "roll3",
- "roll4",
- "roll5",
- "roll6",
- "roll7",
- "roll8"
+ "roll1..9"
],
"q5": [
- "q5.1",
- "q5.2"
+ "q5_1",
+ "q5_2"
],
"q6": [
- "q6.1",
- "q6.2"
+ "q6_1",
+ "q6_2"
],
"q7": [
- "q7.1",
- "q7.2"
+ "q7_1",
+ "q7_2"
],
- "q9": [
- "q9.1",
- "q9.2"
+ "q8": [
+ "q8_1",
+ "q8_2"
],
- "q11": [
- "q8.1",
- "q8.2"
+ "q9": [
+ "q9_1",
+ "q9_2"
]
},
- "Singles": [
- "q1",
- "q2",
- "q3",
- "q4",
- "q10",
- "q11",
- "q12",
- "q13",
- "q14",
- "q15",
- "q16",
- "q17",
- "q18",
- "q19",
- "q20"
- ],
- "QBlocks": {
+ "fieldBlocks": {
"Medium": {
- "qType": "QTYPE_MED",
- "orig": [
- 160,
- 285
- ],
- "bigGaps": [
- 115,
- 11
- ],
- "gaps": [
- 59,
- 46
- ],
- "qNos": [
- [
- [
- "Medium"
- ]
- ]
+ "bubblesGap": 41,
+ "bubbleValues": [
+ "E",
+ "H"
+ ],
+ "direction": "vertical",
+ "fieldLabels": [
+ "Medium"
+ ],
+ "labelsGap": 0,
+ "origin": [
+ 170,
+ 282
]
},
"Roll": {
- "qType": "QTYPE_ROLL",
- "orig": [
- 218,
- 285
- ],
- "bigGaps": [
- 115,
- 11
- ],
- "gaps": [
- 58,
- 46
- ],
- "qNos": [
- [
- [
- "roll0",
- "roll1",
- "roll2",
- "roll3",
- "roll4",
- "roll5",
- "roll6",
- "roll7",
- "roll8"
- ]
- ]
+ "fieldType": "QTYPE_INT",
+ "fieldLabels": [
+ "roll1..9"
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 58,
+ "origin": [
+ 225,
+ 282
]
},
- "Int1": {
- "qType": "QTYPE_INT",
- "orig": [
+ "Int_Block_Q5": {
+ "fieldType": "QTYPE_INT",
+ "fieldLabels": [
+ "q5_1",
+ "q5_2"
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 60,
+ "origin": [
903,
- 285
- ],
- "bigGaps": [
- 128,
- 11
- ],
- "gaps": [
- 59,
- 46
- ],
- "qNos": [
- [
- [
- "q5.1",
- "q5.2"
- ],
- [
- "q6.1",
- "q6.2"
- ],
- [
- "q7.1",
- "q7.2"
- ]
- ]
+ 282
]
},
- "Int2": {
- "qType": "QTYPE_INT",
- "orig": [
- 1418,
- 285
- ],
- "bigGaps": [
- 128,
- 11
- ],
- "gaps": [
- 59,
- 46
- ],
- "qNos": [
- [
- [
- "q8.1",
- "q8.2"
- ],
- [
- "q9.1",
- "q9.2"
- ]
- ]
+ "Int_Block_Q6": {
+ "fieldType": "QTYPE_INT",
+ "fieldLabels": [
+ "q6_1",
+ "q6_2"
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 60,
+ "origin": [
+ 1077,
+ 282
]
},
- "Mcq1": {
- "qType": "QTYPE_MCQ4",
- "orig": [
- 118,
- 857
- ],
- "bigGaps": [
- 115,
- 181
- ],
- "gaps": [
- 59,
- 53
- ],
- "qNos": [
- [
- [
- "q1",
- "q2",
- "q3",
- "q4"
- ],
- [
- "q10",
- "q11",
- "q12",
- "q13"
- ]
- ]
+ "Int_Block_Q7": {
+ "fieldType": "QTYPE_INT",
+ "fieldLabels": [
+ "q7_1",
+ "q7_2"
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 60,
+ "origin": [
+ 1240,
+ 282
+ ]
+ },
+ "Int_Block_Q8": {
+ "fieldType": "QTYPE_INT",
+ "fieldLabels": [
+ "q8_1",
+ "q8_2"
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 57,
+ "origin": [
+ 1410,
+ 282
+ ]
+ },
+ "Int_Block_Q9": {
+ "fieldType": "QTYPE_INT",
+ "fieldLabels": [
+ "q9_1",
+ "q9_2"
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 57,
+ "origin": [
+ 1580,
+ 282
]
},
- "Mcq2": {
- "qType": "QTYPE_MCQ4",
- "orig": [
+ "MCQ_Block_Q1": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q1..4"
+ ],
+ "bubblesGap": 59,
+ "labelsGap": 50,
+ "origin": [
+ 121,
+ 860
+ ]
+ },
+ "MCQ_Block_Q10": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q10..13"
+ ],
+ "bubblesGap": 59,
+ "labelsGap": 50,
+ "origin": [
+ 121,
+ 1195
+ ]
+ },
+ "MCQ_Block_Q14": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q14..16"
+ ],
+ "bubblesGap": 57,
+ "labelsGap": 50,
+ "origin": [
905,
860
- ],
- "bigGaps": [
- 115,
- 180
- ],
- "gaps": [
- 59,
- 53
- ],
- "qNos": [
- [
- [
- "q14",
- "q15",
- "q16"
- ]
- ]
]
},
- "Mcq3": {
- "qType": "QTYPE_MCQ4",
- "orig": [
+ "MCQ_Block_Q17": {
+ "fieldType": "QTYPE_MCQ4",
+ "fieldLabels": [
+ "q17..20"
+ ],
+ "bubblesGap": 57,
+ "labelsGap": 50,
+ "origin": [
905,
- 1198
- ],
- "bigGaps": [
- 115,
- 180
- ],
- "gaps": [
- 59,
- 53
- ],
- "qNos": [
- [
- [
- "q17",
- "q18",
- "q19",
- "q20"
- ]
- ]
+ 1195
]
}
- }
-}
\ No newline at end of file
+ },
+ "preProcessors": [
+ {
+ "name": "CropPage",
+ "options": {
+ "morphKernel": [
+ 10,
+ 10
+ ]
+ }
+ },
+ {
+ "name": "CropOnMarkers",
+ "options": {
+ "relativePath": "omr_marker.jpg",
+ "sheetToMarkerWidthRatio": 17
+ }
+ }
+ ]
+}
diff --git a/samples/sample2/AdrianSample/adrian_omr.png b/samples/sample2/AdrianSample/adrian_omr.png
index dc008145..69a53823 100644
Binary files a/samples/sample2/AdrianSample/adrian_omr.png and b/samples/sample2/AdrianSample/adrian_omr.png differ
diff --git a/samples/sample2/AdrianSample/adrian_omr_2.png b/samples/sample2/AdrianSample/adrian_omr_2.png
new file mode 100644
index 00000000..d8db0994
Binary files /dev/null and b/samples/sample2/AdrianSample/adrian_omr_2.png differ
diff --git a/samples/sample2/config.json b/samples/sample2/config.json
new file mode 100644
index 00000000..a9daf449
--- /dev/null
+++ b/samples/sample2/config.json
@@ -0,0 +1,11 @@
+{
+ "dimensions": {
+ "display_height": 2480,
+ "display_width": 1640,
+ "processing_height": 820,
+ "processing_width": 666
+ },
+ "outputs": {
+ "show_image_level": 5
+ }
+}
diff --git a/samples/sample2/template.json b/samples/sample2/template.json
index e82a9051..41ec9ffa 100644
--- a/samples/sample2/template.json
+++ b/samples/sample2/template.json
@@ -1,46 +1,35 @@
{
- "Dimensions": [
+ "pageDimensions": [
300,
400
],
- "BubbleDimensions": [
+ "bubbleDimensions": [
25,
25
],
- "Concatenations": {},
- "Singles": [
- "q1",
- "q2",
- "q3",
- "q4",
- "q5"
+ "preProcessors": [
+ {
+ "name": "CropPage",
+ "options": {
+ "morphKernel": [
+ 10,
+ 10
+ ]
+ }
+ }
],
- "QBlocks": {
- "MCQBlock1": {
- "qType": "QTYPE_MCQ5",
- "orig": [
+ "fieldBlocks": {
+ "MCQ_Block_1": {
+ "fieldType": "QTYPE_MCQ5",
+ "origin": [
65,
60
],
- "qNos": [
- [
- [
- "q1",
- "q2",
- "q3",
- "q4",
- "q5"
- ]
- ]
- ],
- "gaps": [
- 41,
- 52
+ "fieldLabels": [
+ "q1..5"
],
- "bigGaps": [
- 30,
- 30
- ]
+ "labelsGap": 52,
+ "bubblesGap": 41
}
}
-}
\ No newline at end of file
+}
diff --git a/samples/sample3/README.md b/samples/sample3/README.md
new file mode 100644
index 00000000..d764c8b3
--- /dev/null
+++ b/samples/sample3/README.md
@@ -0,0 +1,31 @@
+## Observation
+The OMR layout is slightly different on colored thick papers vs on xeroxed thin papers. The shift becomes noticible in case of OMR with large number of questions.
+
+We overlapped a colored OMR sheet with a xerox copy of the same OMR sheet(both printed on A4 papers) and observed that there is a great amount of layout sheet as we reach the bottom of the OMR.
+
+Link to an explainer with a real life example: [Google Drive](https://drive.google.com/drive/folders/1GpZTmpEhEjSALJEMjHwDafzWKgEoCTOI?usp=sharing)
+
+## Reasons for shifts in Template layout:
+Listing out a few reasons for the above observation:
+### Printer margin setting
+The margin settings for different printers may be different for the same OMR layout. Thus causing the print to become elongated either horizontally, vertically or both ways.
+
+### The Fan-out effect
+The fan-out effect is usually observed in a sheet fed offset press. Depending on how the papers are made, their dimensions have a tendency to change when they are exposed to moisture or water.
+
+The standard office papers(80 gsm) can easily capture moisture and change shape e.g. in case they get stored for multiple days in a place where the weather is highly humid.
+
+Below are some examples of the GSM ranges:
+
+- 74gsm to 90gsm – This is the basic standard office paper, used in your laser printers.
+- 100gsm to 120gsm – This is stationary paper used for standard letterheads, complimentary slips.
+- 130 to 170gsm – Mostly used for leaflets, posters, single-sided flyers, and brochures.
+
+## Solution
+
+It is recommended to scan each types of prints into different folders and use a separate template.json layout for each of the folders. The same is presented in this sample folder.
+
+## References
+
+- [Paper dimensional stability in sheet-fed offset printing](https://www.diva-portal.org/smash/get/diva2:517895/FULLTEXT01.pdf)
+- An analysis of a few ["Interesting" bubble sheets](https://imgur.com/a/10qwL)
diff --git a/samples/sample3/colored-thick-sheet/rgb-100-gsm.jpg b/samples/sample3/colored-thick-sheet/rgb-100-gsm.jpg
new file mode 100644
index 00000000..4a34731a
Binary files /dev/null and b/samples/sample3/colored-thick-sheet/rgb-100-gsm.jpg differ
diff --git a/samples/sample3/colored-thick-sheet/template.json b/samples/sample3/colored-thick-sheet/template.json
new file mode 100644
index 00000000..743723d6
--- /dev/null
+++ b/samples/sample3/colored-thick-sheet/template.json
@@ -0,0 +1,143 @@
+{
+ "pageDimensions": [
+ 1800,
+ 2400
+ ],
+ "bubbleDimensions": [
+ 23,
+ 20
+ ],
+ "fieldBlocks": {
+ "q01block": {
+ "origin": [
+ 504,
+ 927
+ ],
+ "bubblesGap": 60.35,
+ "labelsGap": 31.75,
+ "fieldLabels": [
+ "q1..10"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q11block": {
+ "origin": [
+ 504,
+ 1242
+ ],
+ "bubblesGap": 60.35,
+ "labelsGap": 31.75,
+ "fieldLabels": [
+ "q11..20"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q21block": {
+ "origin": [
+ 500,
+ 1562
+ ],
+ "bubblesGap": 61.25,
+ "labelsGap": 32.5,
+ "fieldLabels": [
+ "q21..30"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q31block": {
+ "origin": [
+ 500,
+ 1885
+ ],
+ "bubblesGap": 62.25,
+ "labelsGap": 33.5,
+ "fieldLabels": [
+ "q31..40"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q41block": {
+ "origin": [
+ 811,
+ 927
+ ],
+ "bubblesGap": 60.35,
+ "labelsGap": 31.75,
+ "fieldLabels": [
+ "q41..50"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q51block": {
+ "origin": [
+ 811,
+ 1242
+ ],
+ "bubblesGap": 60.35,
+ "labelsGap": 31.75,
+ "fieldLabels": [
+ "q51..60"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q61block": {
+ "origin": [
+ 811,
+ 1562
+ ],
+ "bubblesGap": 61.25,
+ "labelsGap": 32.5,
+ "fieldLabels": [
+ "q61..70"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q71block": {
+ "origin": [
+ 811,
+ 1885
+ ],
+ "bubblesGap": 62.25,
+ "labelsGap": 33.5,
+ "fieldLabels": [
+ "q71..80"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q81block": {
+ "origin": [
+ 1120,
+ 927
+ ],
+ "bubblesGap": 60.35,
+ "labelsGap": 31.75,
+ "fieldLabels": [
+ "q81..90"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q91block": {
+ "origin": [
+ 1120,
+ 1242
+ ],
+ "bubblesGap": 60.35,
+ "labelsGap": 31.75,
+ "fieldLabels": [
+ "q91..100"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ }
+ },
+ "preProcessors": [
+ {
+ "name": "CropPage",
+ "options": {
+ "morphKernel": [
+ 10,
+ 10
+ ]
+ }
+ }
+ ]
+}
diff --git a/samples/sample3/config.json b/samples/sample3/config.json
new file mode 100644
index 00000000..a9daf449
--- /dev/null
+++ b/samples/sample3/config.json
@@ -0,0 +1,11 @@
+{
+ "dimensions": {
+ "display_height": 2480,
+ "display_width": 1640,
+ "processing_height": 820,
+ "processing_width": 666
+ },
+ "outputs": {
+ "show_image_level": 5
+ }
+}
diff --git a/samples/sample3/template.json b/samples/sample3/template.json
deleted file mode 100644
index 2dddf37e..00000000
--- a/samples/sample3/template.json
+++ /dev/null
@@ -1,248 +0,0 @@
-{
- "Dimensions": [
- 1846,
- 1500
- ],
- "BubbleDimensions": [
- 40,
- 40
- ],
- "Options": {
- "Marker": {
- "RelativePath": "omr_marker.jpg",
- "SheetToMarkerWidthRatio": 17
- },
- "OverrideFlags":{
- "noCropping": true
- }
- },
- "Concatenations": {
- "Roll": [
- "Squad",
- "Medium",
- "roll0",
- "roll1",
- "roll2",
- "roll3",
- "roll4",
- "roll5",
- "roll6",
- "roll7",
- "roll8"
- ],
- "q6": [
- "q6.1",
- "q6.2"
- ],
- "q7": [
- "q7.1",
- "q7.2"
- ],
- "q8": [
- "q8.1",
- "q8.2"
- ],
- "q9": [
- "q9.1",
- "q9.2"
- ],
- "q10": [
- "q10.1",
- "q10.2"
- ]
- },
- "Singles": [
- "q1",
- "q2",
- "q3",
- "q4",
- "q5",
- "q11",
- "q12",
- "q13",
- "q14",
- "q15",
- "q16",
- "q17",
- "q18",
- "q19",
- "q20",
- "q21",
- "q22"
- ],
- "QBlocks": {
- "Medium": {
- "qType": "QTYPE_MED",
- "orig": [
- 208,
- 205
- ],
- "bigGaps": [
- 115,
- 11
- ],
- "gaps": [
- 59,
- 46
- ],
- "qNos": [
- [
- [
- "Medium"
- ]
- ]
- ]
- },
- "Roll": {
- "qType": "QTYPE_ROLL",
- "orig": [
- 261,
- 210
- ],
- "bigGaps": [
- 115,
- 11
- ],
- "gaps": [
- 58,
- 46
- ],
- "qNos": [
- [
- [
- "roll0",
- "roll1",
- "roll2",
- "roll3",
- "roll4",
- "roll5",
- "roll6",
- "roll7",
- "roll8"
- ]
- ]
- ]
- },
- "Int1": {
- "qType": "QTYPE_INT",
- "orig": [
- 935,
- 211
- ],
- "bigGaps": [
- 124,
- 11
- ],
- "gaps": [
- 57,
- 46
- ],
- "qNos": [
- [
- [
- "q6.1",
- "q6.2"
- ],
- [
- "q7.1",
- "q7.2"
- ],
- [
- "q8.1",
- "q8.2"
- ],
- [
- "q9.1",
- "q9.2"
- ],
- [
- "q10.1",
- "q10.2"
- ]
- ]
- ]
- },
- "Mcq1": {
- "qType": "QTYPE_MCQ4",
- "orig": [
- 198,
- 826
- ],
- "bigGaps": [
- 115,
- 183
- ],
- "gaps": [
- 93,
- 62
- ],
- "qNos": [
- [
- [
- "q1",
- "q2",
- "q3",
- "q4",
- "q5"
- ]
- ]
- ]
- },
- "Mcq2": {
- "qType": "QTYPE_MCQ4",
- "orig": [
- 833,
- 830
- ],
- "bigGaps": [
- 127,
- 254
- ],
- "gaps": [
- 71,
- 61
- ],
- "qNos": [
- [
- [
- "q11",
- "q12",
- "q13",
- "q14"
- ],
- [
- "q15",
- "q16",
- "q17",
- "q18"
- ]
- ]
- ]
- },
- "Mcq3": {
- "qType": "QTYPE_MCQ4",
- "orig": [
- 1481,
- 830
- ],
- "bigGaps": [
- 115,
- 183
- ],
- "gaps": [
- 73,
- 61
- ],
- "qNos": [
- [
- [
- "q19",
- "q20",
- "q21",
- "q22"
- ]
- ]
- ]
- }
- }
-}
\ No newline at end of file
diff --git a/samples/sample3/xeroxed-thin-sheet/grayscale-80-gsm.jpg b/samples/sample3/xeroxed-thin-sheet/grayscale-80-gsm.jpg
new file mode 100644
index 00000000..3e0e3efa
Binary files /dev/null and b/samples/sample3/xeroxed-thin-sheet/grayscale-80-gsm.jpg differ
diff --git a/samples/sample3/xeroxed-thin-sheet/template.json b/samples/sample3/xeroxed-thin-sheet/template.json
new file mode 100644
index 00000000..8aa4e41c
--- /dev/null
+++ b/samples/sample3/xeroxed-thin-sheet/template.json
@@ -0,0 +1,143 @@
+{
+ "pageDimensions": [
+ 1800,
+ 2400
+ ],
+ "bubbleDimensions": [
+ 23,
+ 20
+ ],
+ "fieldBlocks": {
+ "q01block": {
+ "origin": [
+ 492,
+ 924
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q1..10"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q11block": {
+ "origin": [
+ 492,
+ 1258
+ ],
+ "bubblesGap": 59.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q11..20"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q21block": {
+ "origin": [
+ 492,
+ 1589
+ ],
+ "bubblesGap": 60.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q21..30"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q31block": {
+ "origin": [
+ 487,
+ 1920
+ ],
+ "bubblesGap": 61.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q31..40"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q41block": {
+ "origin": [
+ 807,
+ 924
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q41..50"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q51block": {
+ "origin": [
+ 803,
+ 1258
+ ],
+ "bubblesGap": 59.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q51..60"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q61block": {
+ "origin": [
+ 803,
+ 1589
+ ],
+ "bubblesGap": 60.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q61..70"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q71block": {
+ "origin": [
+ 803,
+ 1920
+ ],
+ "bubblesGap": 60.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q71..80"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q81block": {
+ "origin": [
+ 1115,
+ 924
+ ],
+ "bubblesGap": 58.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q81..90"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ },
+ "q91block": {
+ "origin": [
+ 1115,
+ 1258
+ ],
+ "bubblesGap": 59.75,
+ "labelsGap": 32.65,
+ "fieldLabels": [
+ "q91..100"
+ ],
+ "fieldType": "QTYPE_MCQ4"
+ }
+ },
+ "preProcessors": [
+ {
+ "name": "CropPage",
+ "options": {
+ "morphKernel": [
+ 10,
+ 10
+ ]
+ }
+ }
+ ]
+}
diff --git a/samples/sample4/Class1/sheet1.jpg b/samples/sample4/Class1/sheet1.jpg
deleted file mode 100644
index f3248903..00000000
Binary files a/samples/sample4/Class1/sheet1.jpg and /dev/null differ
diff --git a/samples/sample4/Class2/sheet1.jpg b/samples/sample4/Class2/sheet1.jpg
deleted file mode 100644
index f3248903..00000000
Binary files a/samples/sample4/Class2/sheet1.jpg and /dev/null differ
diff --git a/samples/sample4/IMG_20201116_143512.jpg b/samples/sample4/IMG_20201116_143512.jpg
new file mode 100644
index 00000000..2f591c7a
Binary files /dev/null and b/samples/sample4/IMG_20201116_143512.jpg differ
diff --git a/samples/sample4/IMG_20201116_150717658.jpg b/samples/sample4/IMG_20201116_150717658.jpg
new file mode 100644
index 00000000..592fe425
Binary files /dev/null and b/samples/sample4/IMG_20201116_150717658.jpg differ
diff --git a/samples/sample4/IMG_20201116_150750830.jpg b/samples/sample4/IMG_20201116_150750830.jpg
new file mode 100644
index 00000000..6846ef31
Binary files /dev/null and b/samples/sample4/IMG_20201116_150750830.jpg differ
diff --git a/samples/sample4/config.json b/samples/sample4/config.json
new file mode 100644
index 00000000..df59a460
--- /dev/null
+++ b/samples/sample4/config.json
@@ -0,0 +1,13 @@
+{
+ "dimensions": {
+ "display_width": 1189,
+ "display_height": 1682
+ },
+ "threshold_params": {
+ "MIN_JUMP": 30
+ },
+ "outputs": {
+ "filter_out_multimarked_files": false,
+ "show_image_level": 5
+ }
+}
diff --git a/samples/sample4/evaluation.json b/samples/sample4/evaluation.json
new file mode 100644
index 00000000..50db0fbe
--- /dev/null
+++ b/samples/sample4/evaluation.json
@@ -0,0 +1,34 @@
+{
+ "source_type": "custom",
+ "options": {
+ "questions_in_order": [
+ "q1..11"
+ ],
+ "answers_in_order": [
+ "B",
+ "D",
+ "C",
+ "B",
+ "D",
+ "C",
+ [
+ "B",
+ "C",
+ "BC"
+ ],
+ "A",
+ "C",
+ "D",
+ "C"
+ ],
+ "should_explain_scoring": true,
+ "enable_evaluation_table_to_csv": true
+ },
+ "marking_schemes": {
+ "DEFAULT": {
+ "correct": "3",
+ "incorrect": "-1",
+ "unmarked": "0"
+ }
+ }
+}
diff --git a/samples/sample4/omr_marker.jpg b/samples/sample4/omr_marker.jpg
deleted file mode 100644
index 201dab0a..00000000
Binary files a/samples/sample4/omr_marker.jpg and /dev/null differ
diff --git a/samples/sample4/template.json b/samples/sample4/template.json
index 958d6ef8..0d615b87 100644
--- a/samples/sample4/template.json
+++ b/samples/sample4/template.json
@@ -1,263 +1,45 @@
{
- "Dimensions": [
- 1846,
- 1500
+ "pageDimensions": [
+ 1189,
+ 1682
],
- "BubbleDimensions": [
- 40,
- 40
+ "bubbleDimensions": [
+ 30,
+ 30
],
- "Options": {
- "Marker": {
- "RelativePath": "omr_marker.jpg",
- "SheetToMarkerWidthRatio": 17
+ "preProcessors": [
+ {
+ "name": "GaussianBlur",
+ "options": {
+ "kSize": [
+ 3,
+ 3
+ ],
+ "sigmaX": 0
+ }
},
- "OverrideFlags": {
- "noCropping": true
+ {
+ "name": "CropPage",
+ "options": {
+ "morphKernel": [
+ 10,
+ 10
+ ]
+ }
}
- },
- "Concatenations": {
- "Roll": [
- "Squad",
- "Medium",
- "roll0",
- "roll1",
- "roll2",
- "roll3",
- "roll4",
- "roll5",
- "roll6",
- "roll7",
- "roll8"
- ],
- "q5": [
- "q5.1",
- "q5.2"
- ],
- "q6": [
- "q6.1",
- "q6.2"
- ],
- "q7": [
- "q7.1",
- "q7.2"
- ],
- "q9": [
- "q9.1",
- "q9.2"
- ],
- "q11": [
- "q8.1",
- "q8.2"
- ]
- },
- "Singles": [
- "q1",
- "q2",
- "q3",
- "q4",
- "q10",
- "q11",
- "q12",
- "q13",
- "q14",
- "q15",
- "q16",
- "q17",
- "q18",
- "q19",
- "q20"
],
- "QBlocks": {
- "Medium": {
- "qType": "QTYPE_MED",
- "orig": [
- 160,
- 285
- ],
- "bigGaps": [
- 115,
- 11
- ],
- "gaps": [
- 59,
- 46
- ],
- "qNos": [
- [
- [
- "Medium"
- ]
- ]
- ]
- },
- "Roll": {
- "qType": "QTYPE_ROLL",
- "orig": [
- 218,
- 285
- ],
- "bigGaps": [
- 115,
- 11
- ],
- "gaps": [
- 58,
- 46
- ],
- "qNos": [
- [
- [
- "roll0",
- "roll1",
- "roll2",
- "roll3",
- "roll4",
- "roll5",
- "roll6",
- "roll7",
- "roll8"
- ]
- ]
- ]
- },
- "Int1": {
- "qType": "QTYPE_INT",
- "orig": [
- 903,
- 285
- ],
- "bigGaps": [
- 128,
- 11
- ],
- "gaps": [
- 59,
- 46
- ],
- "qNos": [
- [
- [
- "q5.1",
- "q5.2"
- ],
- [
- "q6.1",
- "q6.2"
- ],
- [
- "q7.1",
- "q7.2"
- ]
- ]
- ]
- },
- "Int2": {
- "qType": "QTYPE_INT",
- "orig": [
- 1418,
- 285
- ],
- "bigGaps": [
- 128,
- 11
- ],
- "gaps": [
- 59,
- 46
- ],
- "qNos": [
- [
- [
- "q8.1",
- "q8.2"
- ],
- [
- "q9.1",
- "q9.2"
- ]
- ]
- ]
- },
- "Mcq1": {
- "qType": "QTYPE_MCQ4",
- "orig": [
- 118,
- 857
- ],
- "bigGaps": [
- 115,
- 181
- ],
- "gaps": [
- 59,
- 53
- ],
- "qNos": [
- [
- [
- "q1",
- "q2",
- "q3",
- "q4"
- ],
- [
- "q10",
- "q11",
- "q12",
- "q13"
- ]
- ]
- ]
- },
- "Mcq2": {
- "qType": "QTYPE_MCQ4",
- "orig": [
- 905,
- 860
- ],
- "bigGaps": [
- 115,
- 180
- ],
- "gaps": [
- 59,
- 53
- ],
- "qNos": [
- [
- [
- "q14",
- "q15",
- "q16"
- ]
- ]
- ]
- },
- "Mcq3": {
- "qType": "QTYPE_MCQ4",
- "orig": [
- 905,
- 1198
- ],
- "bigGaps": [
- 115,
- 180
- ],
- "gaps": [
- 59,
- 53
- ],
- "qNos": [
- [
- [
- "q17",
- "q18",
- "q19",
- "q20"
- ]
- ]
- ]
+ "fieldBlocks": {
+ "MCQBlock1": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 134,
+ 684
+ ],
+ "fieldLabels": [
+ "q1..11"
+ ],
+ "bubblesGap": 79,
+ "labelsGap": 62
}
}
-}
\ No newline at end of file
+}
diff --git a/samples/sample5/README.md b/samples/sample5/README.md
new file mode 100644
index 00000000..b5ed8ccb
--- /dev/null
+++ b/samples/sample5/README.md
@@ -0,0 +1,6 @@
+### OMRChecker Sample
+
+This sample demonstrates multiple things, namely -
+- Running OMRChecker on images scanned using popular document scanning apps
+- Using a common template.json file for sub-folders (e.g. multiple scan batches)
+- Using evaluation.json file with custom marking (without streak-based marking)
diff --git a/samples/sample3/CamScanner/sheet2.jpg b/samples/sample5/ScanBatch1/camscanner-1.jpg
similarity index 100%
rename from samples/sample3/CamScanner/sheet2.jpg
rename to samples/sample5/ScanBatch1/camscanner-1.jpg
diff --git a/samples/sample3/CamScanner/sheet1.jpg b/samples/sample5/ScanBatch2/camscanner-2.jpg
similarity index 100%
rename from samples/sample3/CamScanner/sheet1.jpg
rename to samples/sample5/ScanBatch2/camscanner-2.jpg
diff --git a/samples/sample5/config.json b/samples/sample5/config.json
new file mode 100644
index 00000000..a9daf449
--- /dev/null
+++ b/samples/sample5/config.json
@@ -0,0 +1,11 @@
+{
+ "dimensions": {
+ "display_height": 2480,
+ "display_width": 1640,
+ "processing_height": 820,
+ "processing_width": 666
+ },
+ "outputs": {
+ "show_image_level": 5
+ }
+}
diff --git a/samples/sample5/evaluation.json b/samples/sample5/evaluation.json
new file mode 100644
index 00000000..3332fe6a
--- /dev/null
+++ b/samples/sample5/evaluation.json
@@ -0,0 +1,93 @@
+{
+ "source_type": "custom",
+ "options": {
+ "questions_in_order": [
+ "q1..22"
+ ],
+ "answers_in_order": [
+ "C",
+ "C",
+ "B",
+ "C",
+ "C",
+ [
+ "1",
+ "01"
+ ],
+ "19",
+ "10",
+ "10",
+ "18",
+ "D",
+ "A",
+ "D",
+ "D",
+ "D",
+ "C",
+ "C",
+ "C",
+ "C",
+ "D",
+ "B",
+ "A"
+ ],
+ "should_explain_scoring": true
+ },
+ "marking_schemes": {
+ "DEFAULT": {
+ "correct": "1",
+ "incorrect": "0",
+ "unmarked": "0"
+ },
+ "BOOMERANG_1": {
+ "questions": [
+ "q1..5"
+ ],
+ "marking": {
+ "correct": 4,
+ "incorrect": -1,
+ "unmarked": 0
+ }
+ },
+ "PROXIMITY_1": {
+ "questions": [
+ "q6..10"
+ ],
+ "marking": {
+ "correct": 3,
+ "incorrect": -1,
+ "unmarked": 0
+ }
+ },
+ "FIBONACCI_SECTION_1": {
+ "questions": [
+ "q11..14"
+ ],
+ "marking": {
+ "correct": 2,
+ "incorrect": -1,
+ "unmarked": 0
+ }
+ },
+ "POWER_SECTION_1": {
+ "questions": [
+ "q15..18"
+ ],
+ "marking": {
+ "correct": 1,
+ "incorrect": 0,
+ "unmarked": 0
+ }
+ },
+ "FIBONACCI_SECTION_2": {
+ "questions": [
+ "q19..22"
+ ],
+ "marking": {
+ "correct": 2,
+ "incorrect": -1,
+ "unmarked": 0
+ }
+ }
+ }
+}
diff --git a/samples/sample5/omr_marker.jpg b/samples/sample5/omr_marker.jpg
new file mode 100644
index 00000000..0929feec
Binary files /dev/null and b/samples/sample5/omr_marker.jpg differ
diff --git a/samples/sample5/template.json b/samples/sample5/template.json
new file mode 100644
index 00000000..39f6e83d
--- /dev/null
+++ b/samples/sample5/template.json
@@ -0,0 +1,188 @@
+{
+ "pageDimensions": [
+ 1846,
+ 1500
+ ],
+ "bubbleDimensions": [
+ 40,
+ 40
+ ],
+ "preProcessors": [
+ {
+ "name": "CropOnMarkers",
+ "options": {
+ "relativePath": "omr_marker.jpg",
+ "sheetToMarkerWidthRatio": 17
+ }
+ }
+ ],
+ "customLabels": {
+ "Roll": [
+ "Medium",
+ "roll1..9"
+ ],
+ "q6": [
+ "q6_1",
+ "q6_2"
+ ],
+ "q7": [
+ "q7_1",
+ "q7_2"
+ ],
+ "q8": [
+ "q8_1",
+ "q8_2"
+ ],
+ "q9": [
+ "q9_1",
+ "q9_2"
+ ],
+ "q10": [
+ "q10_1",
+ "q10_2"
+ ]
+ },
+ "fieldBlocks": {
+ "Medium": {
+ "bubbleValues": [
+ "E",
+ "H"
+ ],
+ "direction": "vertical",
+ "origin": [
+ 200,
+ 215
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 0,
+ "fieldLabels": [
+ "Medium"
+ ]
+ },
+ "Roll": {
+ "fieldType": "QTYPE_INT",
+ "origin": [
+ 261,
+ 210
+ ],
+ "bubblesGap": 46.5,
+ "labelsGap": 58,
+ "fieldLabels": [
+ "roll1..9"
+ ]
+ },
+ "Int1": {
+ "fieldType": "QTYPE_INT",
+ "origin": [
+ 935,
+ 211
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 57,
+ "fieldLabels": [
+ "q6_1",
+ "q6_2"
+ ]
+ },
+ "Int2": {
+ "fieldType": "QTYPE_INT",
+ "origin": [
+ 1100,
+ 211
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 57,
+ "fieldLabels": [
+ "q7_1",
+ "q7_2"
+ ]
+ },
+ "Int3": {
+ "fieldType": "QTYPE_INT",
+ "origin": [
+ 1275,
+ 211
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 57,
+ "fieldLabels": [
+ "q8_1",
+ "q8_2"
+ ]
+ },
+ "Int4": {
+ "fieldType": "QTYPE_INT",
+ "origin": [
+ 1449,
+ 211
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 57,
+ "fieldLabels": [
+ "q9_1",
+ "q9_2"
+ ]
+ },
+ "Int5": {
+ "fieldType": "QTYPE_INT",
+ "origin": [
+ 1620,
+ 211
+ ],
+ "bubblesGap": 46,
+ "labelsGap": 57,
+ "fieldLabels": [
+ "q10_1",
+ "q10_2"
+ ]
+ },
+ "Mcq1": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 198,
+ 826
+ ],
+ "bubblesGap": 93,
+ "labelsGap": 62,
+ "fieldLabels": [
+ "q1..5"
+ ]
+ },
+ "Mcq2": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 833,
+ 830
+ ],
+ "bubblesGap": 71,
+ "labelsGap": 61,
+ "fieldLabels": [
+ "q11..14"
+ ]
+ },
+ "Mcq3": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 833,
+ 1270
+ ],
+ "bubblesGap": 71,
+ "labelsGap": 61,
+ "fieldLabels": [
+ "q15..18"
+ ]
+ },
+ "Mcq4": {
+ "fieldType": "QTYPE_MCQ4",
+ "origin": [
+ 1481,
+ 830
+ ],
+ "bubblesGap": 73,
+ "labelsGap": 61,
+ "fieldLabels": [
+ "q19..22"
+ ]
+ }
+ }
+}
diff --git a/samples/sample6/config.json b/samples/sample6/config.json
new file mode 100644
index 00000000..6f39f0ca
--- /dev/null
+++ b/samples/sample6/config.json
@@ -0,0 +1,11 @@
+{
+ "dimensions": {
+ "display_width": 2480,
+ "display_height": 3508,
+ "processing_width": 1653,
+ "processing_height": 2339
+ },
+ "outputs": {
+ "show_image_level": 5
+ }
+}
diff --git a/samples/sample6/doc-scans/sample_roll_01.jpg b/samples/sample6/doc-scans/sample_roll_01.jpg
new file mode 100644
index 00000000..25093a37
Binary files /dev/null and b/samples/sample6/doc-scans/sample_roll_01.jpg differ
diff --git a/samples/sample6/doc-scans/sample_roll_02.jpg b/samples/sample6/doc-scans/sample_roll_02.jpg
new file mode 100644
index 00000000..a81ee4fc
Binary files /dev/null and b/samples/sample6/doc-scans/sample_roll_02.jpg differ
diff --git a/samples/sample6/doc-scans/sample_roll_03.jpg b/samples/sample6/doc-scans/sample_roll_03.jpg
new file mode 100644
index 00000000..94296f4e
Binary files /dev/null and b/samples/sample6/doc-scans/sample_roll_03.jpg differ
diff --git a/samples/sample6/readme.md b/samples/sample6/readme.md
new file mode 100644
index 00000000..2a02ee90
--- /dev/null
+++ b/samples/sample6/readme.md
@@ -0,0 +1,21 @@
+# Demo for feature-based alignment
+
+## Background
+OMR is used to match student roll on final exam scripts. Scripts are scanned using a document scanner and the cover pages are extracted for OMR. Even though a document scanner does not produce any warpped perspective, the alignment is not perfect, causing some rotation and translation in the scans.
+
+The scripts in this sample were specifically selected incorrectly marked scripts to demonstrate how feature-based alignment can correct transformation errors using a reference image. In the actual batch. 156 out of 532 scripts were incorrectly marked. With feature-based alignment, all scripts were correctly marked.
+
+## Usage
+Two template files are given in the sample folder, one with feature-based alignment (template_fb_align), the other without (template_no_fb_align).
+
+## Additional Notes
+
+### Reference Image
+When using a reference image for feature-based alignment, it is better not to have many repeated patterns as it is causes ambiguity when trying to match similar feature points. The bubbles in an OMR form are identical and should not be used for feature-extraction.
+
+Thus, the reference image should be cleared of any bubbles. Forms with lots of text as in this example would be effective.
+
+Note the reference image in this sample was generated from a vector pdf, and not from a scanned blank, producing in a perfectly aligned reference.
+
+### Level adjustment
+The bubbles on the scripts were not shaded dark enough. Thus, a level adjustment was done to bring the black point to 70% to darken the light shading. White point was brought down to 80% to remove the light-grey background in the columns.
diff --git a/samples/sample6/reference.png b/samples/sample6/reference.png
new file mode 100644
index 00000000..5351267f
Binary files /dev/null and b/samples/sample6/reference.png differ
diff --git a/samples/sample6/template.json b/samples/sample6/template.json
new file mode 100644
index 00000000..57605a6c
--- /dev/null
+++ b/samples/sample6/template.json
@@ -0,0 +1,110 @@
+{
+ "pageDimensions": [
+ 2480,
+ 3508
+ ],
+ "bubbleDimensions": [
+ 42,
+ 42
+ ],
+ "preProcessors": [
+ {
+ "name": "Levels",
+ "options": {
+ "low": 0.7,
+ "high": 0.8
+ }
+ },
+ {
+ "name": "GaussianBlur",
+ "options": {
+ "kSize": [
+ 3,
+ 3
+ ],
+ "sigmaX": 0
+ }
+ }
+ ],
+ "customLabels": {
+ "Roll": [
+ "stu",
+ "roll1..7",
+ "check_1",
+ "check_2"
+ ]
+ },
+ "fieldBlocks": {
+ "Check1": {
+ "origin": [
+ 2033,
+ 1290
+ ],
+ "bubblesGap": 50,
+ "labelsGap": 50,
+ "fieldLabels": [
+ "check_1"
+ ],
+ "bubbleValues": [
+ "A",
+ "B",
+ "E",
+ "H",
+ "J",
+ "L",
+ "M"
+ ],
+ "direction": "vertical"
+ },
+ "Check2": {
+ "origin": [
+ 2083,
+ 1290
+ ],
+ "bubblesGap": 50,
+ "labelsGap": 50,
+ "fieldLabels": [
+ "check_2"
+ ],
+ "bubbleValues": [
+ "N",
+ "R",
+ "U",
+ "W",
+ "X",
+ "Y"
+ ],
+ "direction": "vertical"
+ },
+ "Stu": {
+ "origin": [
+ 1636,
+ 1290
+ ],
+ "bubblesGap": 50,
+ "labelsGap": 50,
+ "fieldLabels": [
+ "stu"
+ ],
+ "bubbleValues": [
+ "U",
+ "A",
+ "HT",
+ "GT"
+ ],
+ "direction": "vertical"
+ },
+ "Roll": {
+ "fieldType": "QTYPE_INT",
+ "origin": [
+ 1685,
+ 1290
+ ],
+ "bubblesGap": 50.5,
+ "labelsGap": 50.5,
+ "fieldLabels": [
+ "roll1..7"
+ ]
+ }
+ }
+}
diff --git a/samples/sample6/template_fb_align.json b/samples/sample6/template_fb_align.json
new file mode 100644
index 00000000..7c0c2397
--- /dev/null
+++ b/samples/sample6/template_fb_align.json
@@ -0,0 +1,118 @@
+{
+ "pageDimensions": [
+ 2480,
+ 3508
+ ],
+ "bubbleDimensions": [
+ 42,
+ 42
+ ],
+ "preProcessors": [
+ {
+ "name": "Levels",
+ "options": {
+ "low": 0.7,
+ "high": 0.8
+ }
+ },
+ {
+ "name": "FeatureBasedAlignment",
+ "options": {
+ "reference": "reference.png",
+ "maxFeatures": 1000,
+ "2d": true
+ }
+ },
+ {
+ "name": "GaussianBlur",
+ "options": {
+ "kSize": [
+ 3,
+ 3
+ ],
+ "sigmaX": 0
+ }
+ }
+ ],
+ "customLabels": {
+ "Roll": [
+ "stu",
+ "roll1..7",
+ "check_1",
+ "check_2"
+ ]
+ },
+ "fieldBlocks": {
+ "Check1": {
+ "origin": [
+ 2033,
+ 1290
+ ],
+ "bubblesGap": 50,
+ "labelsGap": 50,
+ "fieldLabels": [
+ "check_1"
+ ],
+ "bubbleValues": [
+ "A",
+ "B",
+ "E",
+ "H",
+ "J",
+ "L",
+ "M"
+ ],
+ "direction": "vertical"
+ },
+ "Check2": {
+ "origin": [
+ 2083,
+ 1290
+ ],
+ "bubblesGap": 50,
+ "labelsGap": 50,
+ "fieldLabels": [
+ "check_2"
+ ],
+ "bubbleValues": [
+ "N",
+ "R",
+ "U",
+ "W",
+ "X",
+ "Y"
+ ],
+ "direction": "vertical"
+ },
+ "Stu": {
+ "origin": [
+ 1636,
+ 1290
+ ],
+ "bubblesGap": 50,
+ "labelsGap": 50,
+ "fieldLabels": [
+ "stu"
+ ],
+ "bubbleValues": [
+ "U",
+ "A",
+ "HT",
+ "GT"
+ ],
+ "direction": "vertical"
+ },
+ "Roll": {
+ "fieldType": "QTYPE_INT",
+ "origin": [
+ 1685,
+ 1290
+ ],
+ "bubblesGap": 50.5,
+ "labelsGap": 50.5,
+ "fieldLabels": [
+ "roll1..7"
+ ]
+ }
+ }
+}
diff --git a/samples/sample6/template_no_fb_align.json b/samples/sample6/template_no_fb_align.json
new file mode 100644
index 00000000..57605a6c
--- /dev/null
+++ b/samples/sample6/template_no_fb_align.json
@@ -0,0 +1,110 @@
+{
+ "pageDimensions": [
+ 2480,
+ 3508
+ ],
+ "bubbleDimensions": [
+ 42,
+ 42
+ ],
+ "preProcessors": [
+ {
+ "name": "Levels",
+ "options": {
+ "low": 0.7,
+ "high": 0.8
+ }
+ },
+ {
+ "name": "GaussianBlur",
+ "options": {
+ "kSize": [
+ 3,
+ 3
+ ],
+ "sigmaX": 0
+ }
+ }
+ ],
+ "customLabels": {
+ "Roll": [
+ "stu",
+ "roll1..7",
+ "check_1",
+ "check_2"
+ ]
+ },
+ "fieldBlocks": {
+ "Check1": {
+ "origin": [
+ 2033,
+ 1290
+ ],
+ "bubblesGap": 50,
+ "labelsGap": 50,
+ "fieldLabels": [
+ "check_1"
+ ],
+ "bubbleValues": [
+ "A",
+ "B",
+ "E",
+ "H",
+ "J",
+ "L",
+ "M"
+ ],
+ "direction": "vertical"
+ },
+ "Check2": {
+ "origin": [
+ 2083,
+ 1290
+ ],
+ "bubblesGap": 50,
+ "labelsGap": 50,
+ "fieldLabels": [
+ "check_2"
+ ],
+ "bubbleValues": [
+ "N",
+ "R",
+ "U",
+ "W",
+ "X",
+ "Y"
+ ],
+ "direction": "vertical"
+ },
+ "Stu": {
+ "origin": [
+ 1636,
+ 1290
+ ],
+ "bubblesGap": 50,
+ "labelsGap": 50,
+ "fieldLabels": [
+ "stu"
+ ],
+ "bubbleValues": [
+ "U",
+ "A",
+ "HT",
+ "GT"
+ ],
+ "direction": "vertical"
+ },
+ "Roll": {
+ "fieldType": "QTYPE_INT",
+ "origin": [
+ 1685,
+ 1290
+ ],
+ "bubblesGap": 50.5,
+ "labelsGap": 50.5,
+ "fieldLabels": [
+ "roll1..7"
+ ]
+ }
+ }
+}
diff --git a/src/__init__.py b/src/__init__.py
new file mode 100644
index 00000000..7fc528b4
--- /dev/null
+++ b/src/__init__.py
@@ -0,0 +1,5 @@
+# https://docs.python.org/3/tutorial/modules.html#:~:text=The%20__init__.py,on%20the%20module%20search%20path.
+from src.logger import logger
+
+# It takes a few seconds for the imports
+logger.info(f"Loading OMRChecker modules...")
diff --git a/src/constants.py b/src/constants.py
new file mode 100644
index 00000000..f1cdc87b
--- /dev/null
+++ b/src/constants.py
@@ -0,0 +1,54 @@
+"""
+
+ OMRChecker
+
+ Author: Udayraj Deshmukh
+ Github: https://github.com/Udayraj123
+
+"""
+from dotmap import DotMap
+
+# Filenames
+TEMPLATE_FILENAME = "template.json"
+EVALUATION_FILENAME = "evaluation.json"
+CONFIG_FILENAME = "config.json"
+
+FIELD_LABEL_NUMBER_REGEX = r"([^\d]+)(\d*)"
+#
+ERROR_CODES = DotMap(
+ {
+ "MULTI_BUBBLE_WARN": 1,
+ "NO_MARKER_ERR": 2,
+ },
+ _dynamic=False,
+)
+
+FIELD_TYPES = {
+ "QTYPE_INT": {
+ "bubbleValues": ["0", "1", "2", "3", "4", "5", "6", "7", "8", "9"],
+ "direction": "vertical",
+ },
+ "QTYPE_INT_FROM_1": {
+ "bubbleValues": ["1", "2", "3", "4", "5", "6", "7", "8", "9", "0"],
+ "direction": "vertical",
+ },
+ "QTYPE_MCQ4": {"bubbleValues": ["A", "B", "C", "D"], "direction": "horizontal"},
+ "QTYPE_MCQ5": {
+ "bubbleValues": ["A", "B", "C", "D", "E"],
+ "direction": "horizontal",
+ },
+ #
+ # You can create and append custom field types here-
+ #
+}
+
+# TODO: move to interaction.py
+TEXT_SIZE = 0.95
+CLR_BLACK = (50, 150, 150)
+CLR_WHITE = (250, 250, 250)
+CLR_GRAY = (130, 130, 130)
+CLR_DARK_GRAY = (100, 100, 100)
+
+# TODO: move to config.json
+GLOBAL_PAGE_THRESHOLD_WHITE = 200
+GLOBAL_PAGE_THRESHOLD_BLACK = 100
diff --git a/src/core.py b/src/core.py
new file mode 100644
index 00000000..d475105f
--- /dev/null
+++ b/src/core.py
@@ -0,0 +1,721 @@
+import os
+from collections import defaultdict
+from typing import Any
+
+import cv2
+import matplotlib.pyplot as plt
+import numpy as np
+
+import src.constants as constants
+from src.logger import logger
+from src.utils.image import CLAHE_HELPER, ImageUtils
+from src.utils.interaction import InteractionUtils
+
+
+class ImageInstanceOps:
+ """Class to hold fine-tuned utilities for a group of images. One instance for each processing directory."""
+
+ save_img_list: Any = defaultdict(list)
+
+ def __init__(self, tuning_config):
+ super().__init__()
+ self.tuning_config = tuning_config
+ self.save_image_level = tuning_config.outputs.save_image_level
+
+ def apply_preprocessors(self, file_path, in_omr, template):
+ tuning_config = self.tuning_config
+ # resize to conform to template
+ in_omr = ImageUtils.resize_util(
+ in_omr,
+ tuning_config.dimensions.processing_width,
+ tuning_config.dimensions.processing_height,
+ )
+
+ # run pre_processors in sequence
+ for pre_processor in template.pre_processors:
+ in_omr = pre_processor.apply_filter(in_omr, file_path)
+ return in_omr
+
+ def read_omr_response(self, template, image, name, save_dir=None):
+ config = self.tuning_config
+ auto_align = config.alignment_params.auto_align
+ try:
+ img = image.copy()
+ # origDim = img.shape[:2]
+ img = ImageUtils.resize_util(
+ img, template.page_dimensions[0], template.page_dimensions[1]
+ )
+ if img.max() > img.min():
+ img = ImageUtils.normalize_util(img)
+ # Processing copies
+ transp_layer = img.copy()
+ final_marked = img.copy()
+
+ morph = img.copy()
+ self.append_save_img(3, morph)
+
+ if auto_align:
+ # Note: clahe is good for morphology, bad for thresholding
+ morph = CLAHE_HELPER.apply(morph)
+ self.append_save_img(3, morph)
+ # Remove shadows further, make columns/boxes darker (less gamma)
+ morph = ImageUtils.adjust_gamma(
+ morph, config.threshold_params.GAMMA_LOW
+ )
+ # TODO: all numbers should come from either constants or config
+ _, morph = cv2.threshold(morph, 220, 220, cv2.THRESH_TRUNC)
+ morph = ImageUtils.normalize_util(morph)
+ self.append_save_img(3, morph)
+ if config.outputs.show_image_level >= 4:
+ InteractionUtils.show("morph1", morph, 0, 1, config)
+
+ # Move them to data class if needed
+ # Overlay Transparencies
+ alpha = 0.65
+ omr_response = {}
+ multi_marked, multi_roll = 0, 0
+
+ # TODO Make this part useful for visualizing status checks
+ # blackVals=[0]
+ # whiteVals=[255]
+
+ if config.outputs.show_image_level >= 5:
+ all_c_box_vals = {"int": [], "mcq": []}
+ # TODO: simplify this logic
+ q_nums = {"int": [], "mcq": []}
+
+ # Find Shifts for the field_blocks --> Before calculating threshold!
+ if auto_align:
+ # print("Begin Alignment")
+ # Open : erode then dilate
+ v_kernel = cv2.getStructuringElement(cv2.MORPH_RECT, (2, 10))
+ morph_v = cv2.morphologyEx(
+ morph, cv2.MORPH_OPEN, v_kernel, iterations=3
+ )
+ _, morph_v = cv2.threshold(morph_v, 200, 200, cv2.THRESH_TRUNC)
+ morph_v = 255 - ImageUtils.normalize_util(morph_v)
+
+ if config.outputs.show_image_level >= 3:
+ InteractionUtils.show(
+ "morphed_vertical", morph_v, 0, 1, config=config
+ )
+
+ # InteractionUtils.show("morph1",morph,0,1,config=config)
+ # InteractionUtils.show("morphed_vertical",morph_v,0,1,config=config)
+
+ self.append_save_img(3, morph_v)
+
+ morph_thr = 60 # for Mobile images, 40 for scanned Images
+ _, morph_v = cv2.threshold(morph_v, morph_thr, 255, cv2.THRESH_BINARY)
+ # kernel best tuned to 5x5 now
+ morph_v = cv2.erode(morph_v, np.ones((5, 5), np.uint8), iterations=2)
+
+ self.append_save_img(3, morph_v)
+ # h_kernel = cv2.getStructuringElement(cv2.MORPH_RECT, (10, 2))
+ # morph_h = cv2.morphologyEx(morph, cv2.MORPH_OPEN, h_kernel, iterations=3)
+ # ret, morph_h = cv2.threshold(morph_h,200,200,cv2.THRESH_TRUNC)
+ # morph_h = 255 - normalize_util(morph_h)
+ # InteractionUtils.show("morph_h",morph_h,0,1,config=config)
+ # _, morph_h = cv2.threshold(morph_h,morph_thr,255,cv2.THRESH_BINARY)
+ # morph_h = cv2.erode(morph_h, np.ones((5,5),np.uint8), iterations = 2)
+ if config.outputs.show_image_level >= 3:
+ InteractionUtils.show(
+ "morph_thr_eroded", morph_v, 0, 1, config=config
+ )
+
+ self.append_save_img(6, morph_v)
+
+ # template relative alignment code
+ for field_block in template.field_blocks:
+ s, d = field_block.origin, field_block.dimensions
+
+ match_col, max_steps, align_stride, thk = map(
+ config.alignment_params.get,
+ [
+ "match_col",
+ "max_steps",
+ "stride",
+ "thickness",
+ ],
+ )
+ shift, steps = 0, 0
+ while steps < max_steps:
+ left_mean = np.mean(
+ morph_v[
+ s[1] : s[1] + d[1],
+ s[0] + shift - thk : -thk + s[0] + shift + match_col,
+ ]
+ )
+ right_mean = np.mean(
+ morph_v[
+ s[1] : s[1] + d[1],
+ s[0]
+ + shift
+ - match_col
+ + d[0]
+ + thk : thk
+ + s[0]
+ + shift
+ + d[0],
+ ]
+ )
+
+ # For demonstration purposes-
+ # if(field_block.name == "int1"):
+ # ret = morph_v.copy()
+ # cv2.rectangle(ret,
+ # (s[0]+shift-thk,s[1]),
+ # (s[0]+shift+thk+d[0],s[1]+d[1]),
+ # constants.CLR_WHITE,
+ # 3)
+ # appendSaveImg(6,ret)
+ # print(shift, left_mean, right_mean)
+ left_shift, right_shift = left_mean > 100, right_mean > 100
+ if left_shift:
+ if right_shift:
+ break
+ else:
+ shift -= align_stride
+ else:
+ if right_shift:
+ shift += align_stride
+ else:
+ break
+ steps += 1
+
+ field_block.shift = shift
+ # print("Aligned field_block: ",field_block.name,"Corrected Shift:",
+ # field_block.shift,", dimensions:", field_block.dimensions,
+ # "origin:", field_block.origin,'\n')
+ # print("End Alignment")
+
+ final_align = None
+ if config.outputs.show_image_level >= 2:
+ initial_align = self.draw_template_layout(img, template, shifted=False)
+ final_align = self.draw_template_layout(
+ img, template, shifted=True, draw_qvals=True
+ )
+ # appendSaveImg(4,mean_vals)
+ self.append_save_img(2, initial_align)
+ self.append_save_img(2, final_align)
+
+ if auto_align:
+ final_align = np.hstack((initial_align, final_align))
+ self.append_save_img(5, img)
+
+ # Get mean bubbleValues n other stats
+ all_q_vals, all_q_strip_arrs, all_q_std_vals = [], [], []
+ total_q_strip_no = 0
+ for field_block in template.field_blocks:
+ box_w, box_h = field_block.bubble_dimensions
+ q_std_vals = []
+ for field_block_bubbles in field_block.traverse_bubbles:
+ q_strip_vals = []
+ for pt in field_block_bubbles:
+ # shifted
+ x, y = (pt.x + field_block.shift, pt.y)
+ rect = [y, y + box_h, x, x + box_w]
+ q_strip_vals.append(
+ cv2.mean(img[rect[0] : rect[1], rect[2] : rect[3]])[0]
+ # detectCross(img, rect) ? 100 : 0
+ )
+ q_std_vals.append(round(np.std(q_strip_vals), 2))
+ all_q_strip_arrs.append(q_strip_vals)
+ # _, _, _ = get_global_threshold(q_strip_vals, "QStrip Plot",
+ # plot_show=False, sort_in_plot=True)
+ # hist = getPlotImg()
+ # InteractionUtils.show("QStrip "+field_block_bubbles[0].field_label, hist, 0, 1,config=config)
+ all_q_vals.extend(q_strip_vals)
+ # print(total_q_strip_no, field_block_bubbles[0].field_label, q_std_vals[len(q_std_vals)-1])
+ total_q_strip_no += 1
+ all_q_std_vals.extend(q_std_vals)
+
+ global_std_thresh, _, _ = self.get_global_threshold(
+ all_q_std_vals
+ ) # , "Q-wise Std-dev Plot", plot_show=True, sort_in_plot=True)
+ # plt.show()
+ # hist = getPlotImg()
+ # InteractionUtils.show("StdHist", hist, 0, 1,config=config)
+
+ # Note: Plotting takes Significant times here --> Change Plotting args
+ # to support show_image_level
+ # , "Mean Intensity Histogram",plot_show=True, sort_in_plot=True)
+ global_thr, _, _ = self.get_global_threshold(all_q_vals, looseness=4)
+
+ logger.info(
+ f"Thresholding: \tglobal_thr: {round(global_thr, 2)} \tglobal_std_THR: {round(global_std_thresh, 2)}\t{'(Looks like a Xeroxed OMR)' if (global_thr == 255) else ''}"
+ )
+ # plt.show()
+ # hist = getPlotImg()
+ # InteractionUtils.show("StdHist", hist, 0, 1,config=config)
+
+ # if(config.outputs.show_image_level>=1):
+ # hist = getPlotImg()
+ # InteractionUtils.show("Hist", hist, 0, 1,config=config)
+ # appendSaveImg(4,hist)
+ # appendSaveImg(5,hist)
+ # appendSaveImg(2,hist)
+
+ per_omr_threshold_avg, total_q_strip_no, total_q_box_no = 0, 0, 0
+ for field_block in template.field_blocks:
+ block_q_strip_no = 1
+ box_w, box_h = field_block.bubble_dimensions
+ shift = field_block.shift
+ s, d = field_block.origin, field_block.dimensions
+ key = field_block.name[:3]
+ # cv2.rectangle(final_marked,(s[0]+shift,s[1]),(s[0]+shift+d[0],
+ # s[1]+d[1]),CLR_BLACK,3)
+ for field_block_bubbles in field_block.traverse_bubbles:
+ # All Black or All White case
+ no_outliers = all_q_std_vals[total_q_strip_no] < global_std_thresh
+ # print(total_q_strip_no, field_block_bubbles[0].field_label,
+ # all_q_std_vals[total_q_strip_no], "no_outliers:", no_outliers)
+ per_q_strip_threshold = self.get_local_threshold(
+ all_q_strip_arrs[total_q_strip_no],
+ global_thr,
+ no_outliers,
+ f"Mean Intensity Histogram for {key}.{field_block_bubbles[0].field_label}.{block_q_strip_no}",
+ config.outputs.show_image_level >= 6,
+ )
+ # print(field_block_bubbles[0].field_label,key,block_q_strip_no, "THR: ",
+ # round(per_q_strip_threshold,2))
+ per_omr_threshold_avg += per_q_strip_threshold
+
+ # Note: Little debugging visualization - view the particular Qstrip
+ # if(
+ # 0
+ # # or "q17" in (field_block_bubbles[0].field_label)
+ # # or (field_block_bubbles[0].field_label+str(block_q_strip_no))=="q15"
+ # ):
+ # st, end = qStrip
+ # InteractionUtils.show("QStrip: "+key+"-"+str(block_q_strip_no),
+ # img[st[1] : end[1], st[0]+shift : end[0]+shift],0,config=config)
+
+ # TODO: get rid of total_q_box_no
+ detected_bubbles = []
+ for bubble in field_block_bubbles:
+ bubble_is_marked = (
+ per_q_strip_threshold > all_q_vals[total_q_box_no]
+ )
+ total_q_box_no += 1
+ if bubble_is_marked:
+ detected_bubbles.append(bubble)
+ x, y, field_value = (
+ bubble.x + field_block.shift,
+ bubble.y,
+ bubble.field_value,
+ )
+ cv2.rectangle(
+ final_marked,
+ (int(x + box_w / 12), int(y + box_h / 12)),
+ (
+ int(x + box_w - box_w / 12),
+ int(y + box_h - box_h / 12),
+ ),
+ constants.CLR_DARK_GRAY,
+ 3,
+ )
+
+ cv2.putText(
+ final_marked,
+ str(field_value),
+ (x, y),
+ cv2.FONT_HERSHEY_SIMPLEX,
+ constants.TEXT_SIZE,
+ (20, 20, 10),
+ int(1 + 3.5 * constants.TEXT_SIZE),
+ )
+ else:
+ cv2.rectangle(
+ final_marked,
+ (int(x + box_w / 10), int(y + box_h / 10)),
+ (
+ int(x + box_w - box_w / 10),
+ int(y + box_h - box_h / 10),
+ ),
+ constants.CLR_GRAY,
+ -1,
+ )
+
+ for bubble in detected_bubbles:
+ field_label, field_value = (
+ bubble.field_label,
+ bubble.field_value,
+ )
+ # Only send rolls multi-marked in the directory
+ multi_marked_local = field_label in omr_response
+ omr_response[field_label] = (
+ (omr_response[field_label] + field_value)
+ if multi_marked_local
+ else field_value
+ )
+ # TODO: generalize this into identifier
+ # multi_roll = multi_marked_local and "Roll" in str(q)
+ multi_marked = multi_marked or multi_marked_local
+
+ if len(detected_bubbles) == 0:
+ field_label = field_block_bubbles[0].field_label
+ omr_response[field_label] = field_block.empty_val
+
+ if config.outputs.show_image_level >= 5:
+ if key in all_c_box_vals:
+ q_nums[key].append(f"{key[:2]}_c{str(block_q_strip_no)}")
+ all_c_box_vals[key].append(
+ all_q_strip_arrs[total_q_strip_no]
+ )
+
+ block_q_strip_no += 1
+ total_q_strip_no += 1
+ # /for field_block
+
+ per_omr_threshold_avg /= total_q_strip_no
+ per_omr_threshold_avg = round(per_omr_threshold_avg, 2)
+ # Translucent
+ cv2.addWeighted(
+ final_marked, alpha, transp_layer, 1 - alpha, 0, final_marked
+ )
+ # Box types
+ if config.outputs.show_image_level >= 6:
+ # plt.draw()
+ f, axes = plt.subplots(len(all_c_box_vals), sharey=True)
+ f.canvas.manager.set_window_title(name)
+ ctr = 0
+ type_name = {
+ "int": "Integer",
+ "mcq": "MCQ",
+ "med": "MED",
+ "rol": "Roll",
+ }
+ for k, boxvals in all_c_box_vals.items():
+ axes[ctr].title.set_text(type_name[k] + " Type")
+ axes[ctr].boxplot(boxvals)
+ # thrline=axes[ctr].axhline(per_omr_threshold_avg,color='red',ls='--')
+ # thrline.set_label("Average THR")
+ axes[ctr].set_ylabel("Intensity")
+ axes[ctr].set_xticklabels(q_nums[k])
+ # axes[ctr].legend()
+ ctr += 1
+ # imshow will do the waiting
+ plt.tight_layout(pad=0.5)
+ plt.show()
+
+ if config.outputs.show_image_level >= 3 and final_align is not None:
+ final_align = ImageUtils.resize_util_h(
+ final_align, int(config.dimensions.display_height)
+ )
+ # [final_align.shape[1],0])
+ InteractionUtils.show(
+ "Template Alignment Adjustment", final_align, 0, 0, config=config
+ )
+
+ if config.outputs.save_detections and save_dir is not None:
+ if multi_roll:
+ save_dir = save_dir.joinpath("_MULTI_")
+ image_path = str(save_dir.joinpath(name))
+ ImageUtils.save_img(image_path, final_marked)
+
+ self.append_save_img(2, final_marked)
+
+ if save_dir is not None:
+ for i in range(config.outputs.save_image_level):
+ self.save_image_stacks(i + 1, name, save_dir)
+
+ return omr_response, final_marked, multi_marked, multi_roll
+
+ except Exception as e:
+ raise e
+
+ @staticmethod
+ def draw_template_layout(img, template, shifted=True, draw_qvals=False, border=-1):
+ img = ImageUtils.resize_util(
+ img, template.page_dimensions[0], template.page_dimensions[1]
+ )
+ final_align = img.copy()
+ for field_block in template.field_blocks:
+ s, d = field_block.origin, field_block.dimensions
+ box_w, box_h = field_block.bubble_dimensions
+ shift = field_block.shift
+ if shifted:
+ cv2.rectangle(
+ final_align,
+ (s[0] + shift, s[1]),
+ (s[0] + shift + d[0], s[1] + d[1]),
+ constants.CLR_BLACK,
+ 3,
+ )
+ else:
+ cv2.rectangle(
+ final_align,
+ (s[0], s[1]),
+ (s[0] + d[0], s[1] + d[1]),
+ constants.CLR_BLACK,
+ 3,
+ )
+ for field_block_bubbles in field_block.traverse_bubbles:
+ for pt in field_block_bubbles:
+ x, y = (pt.x + field_block.shift, pt.y) if shifted else (pt.x, pt.y)
+ cv2.rectangle(
+ final_align,
+ (int(x + box_w / 10), int(y + box_h / 10)),
+ (int(x + box_w - box_w / 10), int(y + box_h - box_h / 10)),
+ constants.CLR_GRAY,
+ border,
+ )
+ if draw_qvals:
+ rect = [y, y + box_h, x, x + box_w]
+ cv2.putText(
+ final_align,
+ f"{int(cv2.mean(img[rect[0] : rect[1], rect[2] : rect[3]])[0])}",
+ (rect[2] + 2, rect[0] + (box_h * 2) // 3),
+ cv2.FONT_HERSHEY_SIMPLEX,
+ 0.6,
+ constants.CLR_BLACK,
+ 2,
+ )
+ if shifted:
+ text_in_px = cv2.getTextSize(
+ field_block.name, cv2.FONT_HERSHEY_SIMPLEX, constants.TEXT_SIZE, 4
+ )
+ cv2.putText(
+ final_align,
+ field_block.name,
+ (int(s[0] + d[0] - text_in_px[0][0]), int(s[1] - text_in_px[0][1])),
+ cv2.FONT_HERSHEY_SIMPLEX,
+ constants.TEXT_SIZE,
+ constants.CLR_BLACK,
+ 4,
+ )
+ return final_align
+
+ def get_global_threshold(
+ self,
+ q_vals_orig,
+ plot_title=None,
+ plot_show=True,
+ sort_in_plot=True,
+ looseness=1,
+ ):
+ """
+ Note: Cannot assume qStrip has only-gray or only-white bg
+ (in which case there is only one jump).
+ So there will be either 1 or 2 jumps.
+ 1 Jump :
+ ......
+ ||||||
+ |||||| <-- risky THR
+ |||||| <-- safe THR
+ ....||||||
+ ||||||||||
+
+ 2 Jumps :
+ ......
+ |||||| <-- wrong THR
+ ....||||||
+ |||||||||| <-- safe THR
+ ..||||||||||
+ ||||||||||||
+
+ The abstract "First LARGE GAP" is perfect for this.
+ Current code is considering ONLY TOP 2 jumps(>= MIN_GAP) to be big,
+ gives the smaller one
+
+ """
+ config = self.tuning_config
+ PAGE_TYPE_FOR_THRESHOLD, MIN_JUMP, JUMP_DELTA = map(
+ config.threshold_params.get,
+ [
+ "PAGE_TYPE_FOR_THRESHOLD",
+ "MIN_JUMP",
+ "JUMP_DELTA",
+ ],
+ )
+
+ global_default_threshold = (
+ constants.GLOBAL_PAGE_THRESHOLD_WHITE
+ if PAGE_TYPE_FOR_THRESHOLD == "white"
+ else constants.GLOBAL_PAGE_THRESHOLD_BLACK
+ )
+
+ # Sort the Q bubbleValues
+ # TODO: Change var name of q_vals
+ q_vals = sorted(q_vals_orig)
+ # Find the FIRST LARGE GAP and set it as threshold:
+ ls = (looseness + 1) // 2
+ l = len(q_vals) - ls
+ max1, thr1 = MIN_JUMP, global_default_threshold
+ for i in range(ls, l):
+ jump = q_vals[i + ls] - q_vals[i - ls]
+ if jump > max1:
+ max1 = jump
+ thr1 = q_vals[i - ls] + jump / 2
+
+ # NOTE: thr2 is deprecated, thus is JUMP_DELTA
+ # Make use of the fact that the JUMP_DELTA(Vertical gap ofc) between
+ # values at detected jumps would be atleast 20
+ max2, thr2 = MIN_JUMP, global_default_threshold
+ # Requires atleast 1 gray box to be present (Roll field will ensure this)
+ for i in range(ls, l):
+ jump = q_vals[i + ls] - q_vals[i - ls]
+ new_thr = q_vals[i - ls] + jump / 2
+ if jump > max2 and abs(thr1 - new_thr) > JUMP_DELTA:
+ max2 = jump
+ thr2 = new_thr
+ # global_thr = min(thr1,thr2)
+ global_thr, j_low, j_high = thr1, thr1 - max1 // 2, thr1 + max1 // 2
+
+ # # For normal images
+ # thresholdRead = 116
+ # if(thr1 > thr2 and thr2 > thresholdRead):
+ # print("Note: taking safer thr line.")
+ # global_thr, j_low, j_high = thr2, thr2 - max2//2, thr2 + max2//2
+
+ if plot_title:
+ _, ax = plt.subplots()
+ ax.bar(range(len(q_vals_orig)), q_vals if sort_in_plot else q_vals_orig)
+ ax.set_title(plot_title)
+ thrline = ax.axhline(global_thr, color="green", ls="--", linewidth=5)
+ thrline.set_label("Global Threshold")
+ thrline = ax.axhline(thr2, color="red", ls=":", linewidth=3)
+ thrline.set_label("THR2 Line")
+ # thrline=ax.axhline(j_low,color='red',ls='-.', linewidth=3)
+ # thrline=ax.axhline(j_high,color='red',ls='-.', linewidth=3)
+ # thrline.set_label("Boundary Line")
+ # ax.set_ylabel("Mean Intensity")
+ ax.set_ylabel("Values")
+ ax.set_xlabel("Position")
+ ax.legend()
+ if plot_show:
+ plt.title(plot_title)
+ plt.show()
+
+ return global_thr, j_low, j_high
+
+ def get_local_threshold(
+ self, q_vals, global_thr, no_outliers, plot_title=None, plot_show=True
+ ):
+ """
+ TODO: Update this documentation too-
+ //No more - Assumption : Colwise background color is uniformly gray or white,
+ but not alternating. In this case there is atmost one jump.
+
+ 0 Jump :
+ <-- safe THR?
+ .......
+ ...|||||||
+ |||||||||| <-- safe THR?
+ // How to decide given range is above or below gray?
+ -> global q_vals shall absolutely help here. Just run same function
+ on total q_vals instead of colwise _//
+ How to decide it is this case of 0 jumps
+
+ 1 Jump :
+ ......
+ ||||||
+ |||||| <-- risky THR
+ |||||| <-- safe THR
+ ....||||||
+ ||||||||||
+
+ """
+ config = self.tuning_config
+ # Sort the Q bubbleValues
+ q_vals = sorted(q_vals)
+
+ # Small no of pts cases:
+ # base case: 1 or 2 pts
+ if len(q_vals) < 3:
+ thr1 = (
+ global_thr
+ if np.max(q_vals) - np.min(q_vals) < config.threshold_params.MIN_GAP
+ else np.mean(q_vals)
+ )
+ else:
+ # qmin, qmax, qmean, qstd = round(np.min(q_vals),2), round(np.max(q_vals),2),
+ # round(np.mean(q_vals),2), round(np.std(q_vals),2)
+ # GVals = [round(abs(q-qmean),2) for q in q_vals]
+ # gmean, gstd = round(np.mean(GVals),2), round(np.std(GVals),2)
+ # # DISCRETION: Pretty critical factor in reading response
+ # # Doesn't work well for small number of values.
+ # DISCRETION = 2.7 # 2.59 was closest hit, 3.0 is too far
+ # L2MaxGap = round(max([abs(g-gmean) for g in GVals]),2)
+ # if(L2MaxGap > DISCRETION*gstd):
+ # no_outliers = False
+
+ # # ^Stackoverflow method
+ # print(field_label, no_outliers,"qstd",round(np.std(q_vals),2), "gstd", gstd,
+ # "Gaps in gvals",sorted([round(abs(g-gmean),2) for g in GVals],reverse=True),
+ # '\t',round(DISCRETION*gstd,2), L2MaxGap)
+
+ # else:
+ # Find the LARGEST GAP and set it as threshold: //(FIRST LARGE GAP)
+ l = len(q_vals) - 1
+ max1, thr1 = config.threshold_params.MIN_JUMP, 255
+ for i in range(1, l):
+ jump = q_vals[i + 1] - q_vals[i - 1]
+ if jump > max1:
+ max1 = jump
+ thr1 = q_vals[i - 1] + jump / 2
+ # print(field_label,q_vals,max1)
+
+ confident_jump = (
+ config.threshold_params.MIN_JUMP
+ + config.threshold_params.CONFIDENT_SURPLUS
+ )
+ # If not confident, then only take help of global_thr
+ if max1 < confident_jump:
+ if no_outliers:
+ # All Black or All White case
+ thr1 = global_thr
+ else:
+ # TODO: Low confidence parameters here
+ pass
+
+ # if(thr1 == 255):
+ # print("Warning: threshold is unexpectedly 255! (Outlier Delta issue?)",plot_title)
+
+ # Make a common plot function to show local and global thresholds
+ if plot_show and plot_title is not None:
+ _, ax = plt.subplots()
+ ax.bar(range(len(q_vals)), q_vals)
+ thrline = ax.axhline(thr1, color="green", ls=("-."), linewidth=3)
+ thrline.set_label("Local Threshold")
+ thrline = ax.axhline(global_thr, color="red", ls=":", linewidth=5)
+ thrline.set_label("Global Threshold")
+ ax.set_title(plot_title)
+ ax.set_ylabel("Bubble Mean Intensity")
+ ax.set_xlabel("Bubble Number(sorted)")
+ ax.legend()
+ # TODO append QStrip to this plot-
+ # appendSaveImg(6,getPlotImg())
+ if plot_show:
+ plt.show()
+ return thr1
+
+ def append_save_img(self, key, img):
+ if self.save_image_level >= int(key):
+ self.save_img_list[key].append(img.copy())
+
+ def save_image_stacks(self, key, filename, save_dir):
+ config = self.tuning_config
+ if self.save_image_level >= int(key) and self.save_img_list[key] != []:
+ name = os.path.splitext(filename)[0]
+ result = np.hstack(
+ tuple(
+ [
+ ImageUtils.resize_util_h(img, config.dimensions.display_height)
+ for img in self.save_img_list[key]
+ ]
+ )
+ )
+ result = ImageUtils.resize_util(
+ result,
+ min(
+ len(self.save_img_list[key]) * config.dimensions.display_width // 3,
+ int(config.dimensions.display_width * 2.5),
+ ),
+ )
+ ImageUtils.save_img(f"{save_dir}stack/{name}_{str(key)}_stack.jpg", result)
+
+ def reset_all_save_img(self):
+ for i in range(self.save_image_level):
+ self.save_img_list[i + 1] = []
diff --git a/src/defaults/__init__.py b/src/defaults/__init__.py
new file mode 100644
index 00000000..62ed2e6f
--- /dev/null
+++ b/src/defaults/__init__.py
@@ -0,0 +1,5 @@
+# https://docs.python.org/3/tutorial/modules.html#:~:text=The%20__init__.py,on%20the%20module%20search%20path.
+# Use all imports relative to root directory
+# (https://chrisyeh96.github.io/2017/08/08/definitive-guide-python-imports.html)
+from src.defaults.config import * # NOQA
+from src.defaults.template import * # NOQA
diff --git a/src/defaults/config.py b/src/defaults/config.py
new file mode 100644
index 00000000..1c6cad3f
--- /dev/null
+++ b/src/defaults/config.py
@@ -0,0 +1,35 @@
+from dotmap import DotMap
+
+CONFIG_DEFAULTS = DotMap(
+ {
+ "dimensions": {
+ "display_height": 2480,
+ "display_width": 1640,
+ "processing_height": 820,
+ "processing_width": 666,
+ },
+ "threshold_params": {
+ "GAMMA_LOW": 0.7,
+ "MIN_GAP": 30,
+ "MIN_JUMP": 25,
+ "CONFIDENT_SURPLUS": 5,
+ "JUMP_DELTA": 30,
+ "PAGE_TYPE_FOR_THRESHOLD": "white",
+ },
+ "alignment_params": {
+ # Note: 'auto_align' enables automatic template alignment, use if the scans show slight misalignments.
+ "auto_align": False,
+ "match_col": 5,
+ "max_steps": 20,
+ "stride": 1,
+ "thickness": 3,
+ },
+ "outputs": {
+ "show_image_level": 0,
+ "save_image_level": 0,
+ "save_detections": True,
+ "filter_out_multimarked_files": False,
+ },
+ },
+ _dynamic=False,
+)
diff --git a/src/defaults/template.py b/src/defaults/template.py
new file mode 100644
index 00000000..d0a2a831
--- /dev/null
+++ b/src/defaults/template.py
@@ -0,0 +1,6 @@
+TEMPLATE_DEFAULTS = {
+ "preProcessors": [],
+ "emptyValue": "",
+ "customLabels": {},
+ "outputColumns": [],
+}
diff --git a/src/entry.py b/src/entry.py
new file mode 100644
index 00000000..5cfcfd95
--- /dev/null
+++ b/src/entry.py
@@ -0,0 +1,370 @@
+"""
+
+ OMRChecker
+
+ Author: Udayraj Deshmukh
+ Github: https://github.com/Udayraj123
+
+"""
+import os
+from csv import QUOTE_NONNUMERIC
+from pathlib import Path
+from time import time
+
+import cv2
+import pandas as pd
+from rich.table import Table
+
+from src import constants
+from src.defaults import CONFIG_DEFAULTS
+from src.evaluation import EvaluationConfig, evaluate_concatenated_response
+from src.logger import console, logger
+from src.template import Template
+from src.utils.file import Paths, setup_dirs_for_paths, setup_outputs_for_template
+from src.utils.image import ImageUtils
+from src.utils.interaction import InteractionUtils, Stats
+from src.utils.parsing import get_concatenated_response, open_config_with_defaults
+
+# Load processors
+STATS = Stats()
+
+
+def entry_point(input_dir, args):
+ if not os.path.exists(input_dir):
+ raise Exception(f"Given input directory does not exist: '{input_dir}'")
+ curr_dir = input_dir
+ return process_dir(input_dir, curr_dir, args)
+
+
+def print_config_summary(
+ curr_dir,
+ omr_files,
+ template,
+ tuning_config,
+ local_config_path,
+ evaluation_config,
+ args,
+):
+ logger.info("")
+ table = Table(title="Current Configurations", show_header=False, show_lines=False)
+ table.add_column("Key", style="cyan", no_wrap=True)
+ table.add_column("Value", style="magenta")
+ table.add_row("Directory Path", f"{curr_dir}")
+ table.add_row("Count of Images", f"{len(omr_files)}")
+ table.add_row("Set Layout Mode ", "ON" if args["setLayout"] else "OFF")
+ table.add_row(
+ "Markers Detection",
+ "ON" if "CropOnMarkers" in template.pre_processors else "OFF",
+ )
+ table.add_row("Auto Alignment", f"{tuning_config.alignment_params.auto_align}")
+ table.add_row("Detected Template Path", f"{template}")
+ if local_config_path:
+ table.add_row("Detected Local Config", f"{local_config_path}")
+ if evaluation_config:
+ table.add_row("Detected Evaluation Config", f"{evaluation_config}")
+
+ table.add_row(
+ "Detected pre-processors",
+ f"{[pp.__class__.__name__ for pp in template.pre_processors]}",
+ )
+ console.print(table, justify="center")
+
+
+def process_dir(
+ root_dir,
+ curr_dir,
+ args,
+ template=None,
+ tuning_config=CONFIG_DEFAULTS,
+ evaluation_config=None,
+):
+ # Update local tuning_config (in current recursion stack)
+ local_config_path = curr_dir.joinpath(constants.CONFIG_FILENAME)
+ if os.path.exists(local_config_path):
+ tuning_config = open_config_with_defaults(local_config_path)
+
+ # Update local template (in current recursion stack)
+ local_template_path = curr_dir.joinpath(constants.TEMPLATE_FILENAME)
+ local_template_exists = os.path.exists(local_template_path)
+ if local_template_exists:
+ template = Template(
+ local_template_path,
+ tuning_config,
+ )
+ # Look for subdirectories for processing
+ subdirs = [d for d in curr_dir.iterdir() if d.is_dir()]
+
+ output_dir = Path(args["output_dir"], curr_dir.relative_to(root_dir))
+ paths = Paths(output_dir)
+
+ # look for images in current dir to process
+ exts = ("*.[pP][nN][gG]", "*.[jJ][pP][gG]", "*.[jJ][pP][eE][gG]")
+ omr_files = sorted([f for ext in exts for f in curr_dir.glob(ext)])
+
+ # Exclude images (take union over all pre_processors)
+ excluded_files = []
+ if template:
+ for pp in template.pre_processors:
+ excluded_files.extend(Path(p) for p in pp.exclude_files())
+
+ local_evaluation_path = curr_dir.joinpath(constants.EVALUATION_FILENAME)
+ if not args["setLayout"] and os.path.exists(local_evaluation_path):
+ if not local_template_exists:
+ logger.warning(
+ f"Found an evaluation file without a parent template file: {local_evaluation_path}"
+ )
+ evaluation_config = EvaluationConfig(
+ curr_dir,
+ local_evaluation_path,
+ template,
+ tuning_config,
+ )
+
+ excluded_files.extend(
+ Path(exclude_file) for exclude_file in evaluation_config.get_exclude_files()
+ )
+
+ omr_files = [f for f in omr_files if f not in excluded_files]
+
+ if omr_files:
+ if not template:
+ logger.error(
+ f"Found images, but no template in the directory tree \
+ of '{curr_dir}'. \nPlace {constants.TEMPLATE_FILENAME} in the \
+ appropriate directory."
+ )
+ raise Exception(
+ f"No template file found in the directory tree of {curr_dir}"
+ )
+
+ setup_dirs_for_paths(paths)
+ outputs_namespace = setup_outputs_for_template(paths, template)
+
+ print_config_summary(
+ curr_dir,
+ omr_files,
+ template,
+ tuning_config,
+ local_config_path,
+ evaluation_config,
+ args,
+ )
+ if args["setLayout"]:
+ show_template_layouts(omr_files, template, tuning_config)
+ else:
+ process_files(
+ omr_files,
+ template,
+ tuning_config,
+ evaluation_config,
+ outputs_namespace,
+ )
+
+ elif not subdirs:
+ # Each subdirectory should have images or should be non-leaf
+ logger.info(
+ f"No valid images or sub-folders found in {curr_dir}.\
+ Empty directories not allowed."
+ )
+
+ # recursively process sub-folders
+ for d in subdirs:
+ process_dir(
+ root_dir,
+ d,
+ args,
+ template,
+ tuning_config,
+ evaluation_config,
+ )
+
+
+def show_template_layouts(omr_files, template, tuning_config):
+ for file_path in omr_files:
+ file_name = file_path.name
+ file_path = str(file_path)
+ in_omr = cv2.imread(file_path, cv2.IMREAD_GRAYSCALE)
+ in_omr = template.image_instance_ops.apply_preprocessors(
+ file_path, in_omr, template
+ )
+ template_layout = template.image_instance_ops.draw_template_layout(
+ in_omr, template, shifted=False, border=2
+ )
+ InteractionUtils.show(
+ f"Template Layout: {file_name}", template_layout, 1, 1, config=tuning_config
+ )
+
+
+def process_files(
+ omr_files,
+ template,
+ tuning_config,
+ evaluation_config,
+ outputs_namespace,
+):
+ start_time = int(time())
+ files_counter = 0
+ STATS.files_not_moved = 0
+
+ for file_path in omr_files:
+ files_counter += 1
+ file_name = file_path.name
+
+ in_omr = cv2.imread(str(file_path), cv2.IMREAD_GRAYSCALE)
+
+ logger.info("")
+ logger.info(
+ f"({files_counter}) Opening image: \t'{file_path}'\tResolution: {in_omr.shape}"
+ )
+
+ template.image_instance_ops.reset_all_save_img()
+
+ template.image_instance_ops.append_save_img(1, in_omr)
+
+ in_omr = template.image_instance_ops.apply_preprocessors(
+ file_path, in_omr, template
+ )
+
+ if in_omr is None:
+ # Error OMR case
+ new_file_path = outputs_namespace.paths.errors_dir.joinpath(file_name)
+ outputs_namespace.OUTPUT_SET.append(
+ [file_name] + outputs_namespace.empty_resp
+ )
+ if check_and_move(
+ constants.ERROR_CODES.NO_MARKER_ERR, file_path, new_file_path
+ ):
+ err_line = [
+ file_name,
+ file_path,
+ new_file_path,
+ "NA",
+ ] + outputs_namespace.empty_resp
+ pd.DataFrame(err_line, dtype=str).T.to_csv(
+ outputs_namespace.files_obj["Errors"],
+ mode="a",
+ quoting=QUOTE_NONNUMERIC,
+ header=False,
+ index=False,
+ )
+ continue
+
+ # uniquify
+ file_id = str(file_name)
+ save_dir = outputs_namespace.paths.save_marked_dir
+ (
+ response_dict,
+ final_marked,
+ multi_marked,
+ _,
+ ) = template.image_instance_ops.read_omr_response(
+ template, image=in_omr, name=file_id, save_dir=save_dir
+ )
+
+ # TODO: move inner try catch here
+ # concatenate roll nos, set unmarked responses, etc
+ omr_response = get_concatenated_response(response_dict, template)
+
+ if (
+ evaluation_config is None
+ or not evaluation_config.get_should_explain_scoring()
+ ):
+ logger.info(f"Read Response: \n{omr_response}")
+
+ score = 0
+ if evaluation_config is not None:
+ score = evaluate_concatenated_response(
+ omr_response, evaluation_config, file_path, outputs_namespace.paths.evaluation_dir
+ )
+ logger.info(
+ f"(/{files_counter}) Graded with score: {round(score, 2)}\t for file: '{file_id}'"
+ )
+ else:
+ logger.info(f"(/{files_counter}) Processed file: '{file_id}'")
+
+ if tuning_config.outputs.show_image_level >= 2:
+ InteractionUtils.show(
+ f"Final Marked Bubbles : '{file_id}'",
+ ImageUtils.resize_util_h(
+ final_marked, int(tuning_config.dimensions.display_height * 1.3)
+ ),
+ 1,
+ 1,
+ config=tuning_config,
+ )
+
+ resp_array = []
+ for k in template.output_columns:
+ resp_array.append(omr_response[k])
+
+ outputs_namespace.OUTPUT_SET.append([file_name] + resp_array)
+
+ if multi_marked == 0 or not tuning_config.outputs.filter_out_multimarked_files:
+ STATS.files_not_moved += 1
+ new_file_path = save_dir.joinpath(file_id)
+ # Enter into Results sheet-
+ results_line = [file_name, file_path, new_file_path, score] + resp_array
+ # Write/Append to results_line file(opened in append mode)
+ pd.DataFrame(results_line, dtype=str).T.to_csv(
+ outputs_namespace.files_obj["Results"],
+ mode="a",
+ quoting=QUOTE_NONNUMERIC,
+ header=False,
+ index=False,
+ )
+ else:
+ # multi_marked file
+ logger.info(f"[{files_counter}] Found multi-marked file: '{file_id}'")
+ new_file_path = outputs_namespace.paths.multi_marked_dir.joinpath(file_name)
+ if check_and_move(
+ constants.ERROR_CODES.MULTI_BUBBLE_WARN, file_path, new_file_path
+ ):
+ mm_line = [file_name, file_path, new_file_path, "NA"] + resp_array
+ pd.DataFrame(mm_line, dtype=str).T.to_csv(
+ outputs_namespace.files_obj["MultiMarked"],
+ mode="a",
+ quoting=QUOTE_NONNUMERIC,
+ header=False,
+ index=False,
+ )
+ # else:
+ # TODO: Add appropriate record handling here
+ # pass
+
+ print_stats(start_time, files_counter, tuning_config)
+
+
+def check_and_move(error_code, file_path, filepath2):
+ # TODO: fix file movement into error/multimarked/invalid etc again
+ STATS.files_not_moved += 1
+ return True
+
+
+def print_stats(start_time, files_counter, tuning_config):
+ time_checking = max(1, round(time() - start_time, 2))
+ log = logger.info
+ log("")
+ log(f"{'Total file(s) moved': <27}: {STATS.files_moved}")
+ log(f"{'Total file(s) not moved': <27}: {STATS.files_not_moved}")
+ log("--------------------------------")
+ log(
+ f"{'Total file(s) processed': <27}: {files_counter} ({'Sum Tallied!' if files_counter == (STATS.files_moved + STATS.files_not_moved) else 'Not Tallying!'})"
+ )
+
+ if tuning_config.outputs.show_image_level <= 0:
+ log(
+ f"\nFinished Checking {files_counter} file(s) in {round(time_checking, 1)} seconds i.e. ~{round(time_checking / 60, 1)} minute(s)."
+ )
+ log(
+ f"{'OMR Processing Rate': <27}: \t ~ {round(time_checking / files_counter, 2)} seconds/OMR"
+ )
+ log(
+ f"{'OMR Processing Speed': <27}: \t ~ {round((files_counter * 60) / time_checking, 2)} OMRs/minute"
+ )
+ else:
+ log(f"\n{'Total script time': <27}: {time_checking} seconds")
+
+ if tuning_config.outputs.show_image_level <= 1:
+ log(
+ "\nTip: To see some awesome visuals, open config.json and increase 'show_image_level'"
+ )
diff --git a/src/evaluation.py b/src/evaluation.py
new file mode 100644
index 00000000..0b567169
--- /dev/null
+++ b/src/evaluation.py
@@ -0,0 +1,546 @@
+import ast
+import os
+import re
+from copy import deepcopy
+from csv import QUOTE_NONNUMERIC
+
+import cv2
+import pandas as pd
+from rich.table import Table
+
+from src.logger import console, logger
+from src.schemas.constants import (
+ BONUS_SECTION_PREFIX,
+ DEFAULT_SECTION_KEY,
+ MARKING_VERDICT_TYPES,
+)
+from src.utils.parsing import (
+ get_concatenated_response,
+ open_evaluation_with_validation,
+ parse_fields,
+ parse_float_or_fraction,
+)
+
+
+class AnswerMatcher:
+ def __init__(self, answer_item, section_marking_scheme):
+ self.section_marking_scheme = section_marking_scheme
+ self.answer_item = answer_item
+ self.answer_type = self.validate_and_get_answer_type(answer_item)
+ self.set_defaults_from_scheme(section_marking_scheme)
+
+ @staticmethod
+ def is_a_marking_score(answer_element):
+ # Note: strict type checking is already done at schema validation level,
+ # Here we focus on overall struct type
+ return type(answer_element) == str or type(answer_element) == int
+
+ @staticmethod
+ def is_standard_answer(answer_element):
+ return type(answer_element) == str and len(answer_element) >= 1
+
+ def validate_and_get_answer_type(self, answer_item):
+ if self.is_standard_answer(answer_item):
+ return "standard"
+ elif type(answer_item) == list:
+ if (
+ # Array of answer elements: ['A', 'B', 'AB']
+ len(answer_item) >= 2
+ and all(
+ self.is_standard_answer(answers_or_score)
+ for answers_or_score in answer_item
+ )
+ ):
+ return "multiple-correct"
+ elif (
+ # Array of two-tuples: [['A', 1], ['B', 1], ['C', 3], ['AB', 2]]
+ len(answer_item) >= 1
+ and all(
+ type(answer_and_score) == list and len(answer_and_score) == 2
+ for answer_and_score in answer_item
+ )
+ and all(
+ self.is_standard_answer(allowed_answer)
+ and self.is_a_marking_score(answer_score)
+ for allowed_answer, answer_score in answer_item
+ )
+ ):
+ return "multiple-correct-weighted"
+
+ logger.critical(
+ f"Unable to determine answer type for answer item: {answer_item}"
+ )
+ raise Exception("Unable to determine answer type")
+
+ def set_defaults_from_scheme(self, section_marking_scheme):
+ answer_type = self.answer_type
+ self.empty_val = section_marking_scheme.empty_val
+ answer_item = self.answer_item
+ self.marking = deepcopy(section_marking_scheme.marking)
+ # TODO: reuse part of parse_scheme_marking here -
+ if answer_type == "standard":
+ # no local overrides
+ pass
+ elif answer_type == "multiple-correct":
+ # override marking scheme scores for each allowed answer
+ for allowed_answer in answer_item:
+ self.marking[f"correct-{allowed_answer}"] = self.marking["correct"]
+ elif answer_type == "multiple-correct-weighted":
+ # Note: No override using marking scheme as answer scores are provided in answer_item
+ for allowed_answer, answer_score in answer_item:
+ self.marking[f"correct-{allowed_answer}"] = parse_float_or_fraction(
+ answer_score
+ )
+
+ def get_marking_scheme(self):
+ return self.section_marking_scheme
+
+ def get_section_explanation(self):
+ answer_type = self.answer_type
+ if answer_type in ["standard", "multiple-correct"]:
+ return self.section_marking_scheme.section_key
+ elif answer_type == "multiple-correct-weighted":
+ return f"Custom: {self.marking}"
+
+ def get_verdict_marking(self, marked_answer):
+ answer_type = self.answer_type
+ question_verdict = "incorrect"
+ if answer_type == "standard":
+ question_verdict = self.get_standard_verdict(marked_answer)
+ elif answer_type == "multiple-correct":
+ question_verdict = self.get_multiple_correct_verdict(marked_answer)
+ elif answer_type == "multiple-correct-weighted":
+ question_verdict = self.get_multiple_correct_weighted_verdict(marked_answer)
+ return question_verdict, self.marking[question_verdict]
+
+ def get_standard_verdict(self, marked_answer):
+ allowed_answer = self.answer_item
+ if marked_answer == self.empty_val:
+ return "unmarked"
+ elif marked_answer == allowed_answer:
+ return "correct"
+ else:
+ return "incorrect"
+
+ def get_multiple_correct_verdict(self, marked_answer):
+ allowed_answers = self.answer_item
+ if marked_answer == self.empty_val:
+ return "unmarked"
+ elif marked_answer in allowed_answers:
+ return f"correct-{marked_answer}"
+ else:
+ return "incorrect"
+
+ def get_multiple_correct_weighted_verdict(self, marked_answer):
+ allowed_answers = [
+ allowed_answer for allowed_answer, _answer_score in self.answer_item
+ ]
+ if marked_answer == self.empty_val:
+ return "unmarked"
+ elif marked_answer in allowed_answers:
+ return f"correct-{marked_answer}"
+ else:
+ return "incorrect"
+
+ def __str__(self):
+ return f"{self.answer_item}"
+
+
+class SectionMarkingScheme:
+ def __init__(self, section_key, section_scheme, empty_val):
+ # TODO: get local empty_val from qblock
+ self.empty_val = empty_val
+ self.section_key = section_key
+ # DEFAULT marking scheme follows a shorthand
+ if section_key == DEFAULT_SECTION_KEY:
+ self.questions = None
+ self.marking = self.parse_scheme_marking(section_scheme)
+ else:
+ self.questions = parse_fields(section_key, section_scheme["questions"])
+ self.marking = self.parse_scheme_marking(section_scheme["marking"])
+
+ def __str__(self):
+ return self.section_key
+
+ def parse_scheme_marking(self, marking):
+ parsed_marking = {}
+ for verdict_type in MARKING_VERDICT_TYPES:
+ verdict_marking = parse_float_or_fraction(marking[verdict_type])
+ if (
+ verdict_marking > 0
+ and verdict_type == "incorrect"
+ and not self.section_key.startswith(BONUS_SECTION_PREFIX)
+ ):
+ logger.warning(
+ f"Found positive marks({round(verdict_marking, 2)}) for incorrect answer in the schema '{self.section_key}'. For Bonus sections, add a prefix 'BONUS_' to them."
+ )
+ parsed_marking[verdict_type] = verdict_marking
+
+ return parsed_marking
+
+ def match_answer(self, marked_answer, answer_matcher):
+ question_verdict, verdict_marking = answer_matcher.get_verdict_marking(
+ marked_answer
+ )
+
+ return verdict_marking, question_verdict
+
+
+class EvaluationConfig:
+ """Note: this instance will be reused for multiple omr sheets"""
+
+ def __init__(self, curr_dir, evaluation_path, template, tuning_config):
+ self.path = evaluation_path
+ evaluation_json = open_evaluation_with_validation(evaluation_path)
+ options, marking_schemes, source_type = map(
+ evaluation_json.get, ["options", "marking_schemes", "source_type"]
+ )
+ self.should_explain_scoring = options.get("should_explain_scoring", False)
+ self.has_non_default_section = False
+ self.exclude_files = []
+ self.enable_evaluation_table_to_csv = options.get(
+ "enable_evaluation_table_to_csv", False
+ )
+
+ if source_type == "csv":
+ csv_path = curr_dir.joinpath(options["answer_key_csv_path"])
+ if not os.path.exists(csv_path):
+ logger.warning(f"Answer key csv does not exist at: '{csv_path}'.")
+
+ answer_key_image_path = options.get("answer_key_image_path", None)
+ if os.path.exists(csv_path):
+ # TODO: CSV parsing/validation for each row with a (qNo,